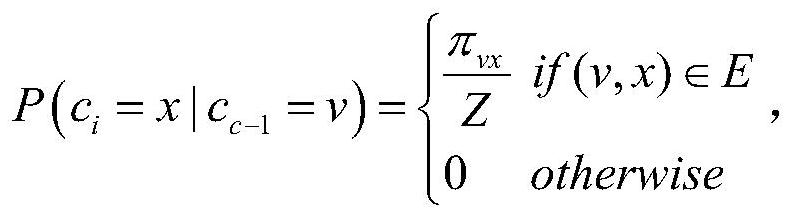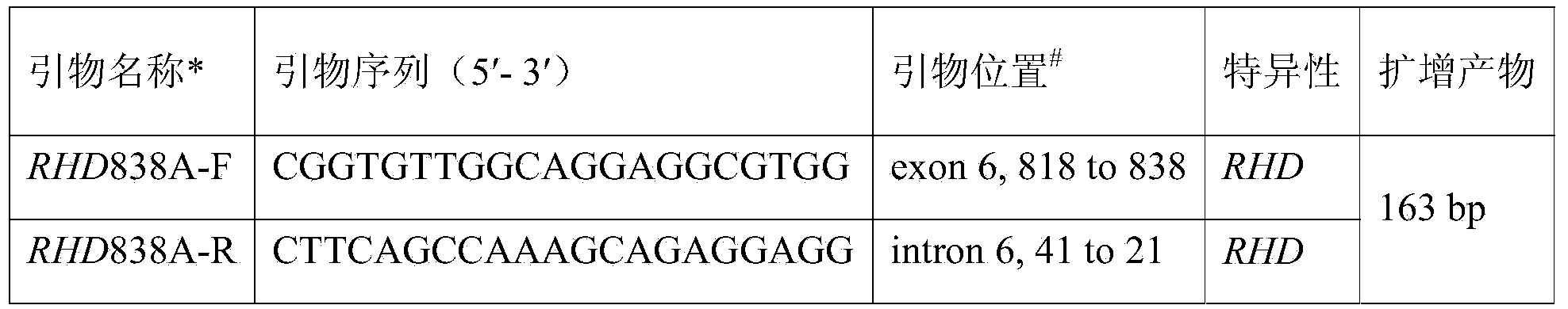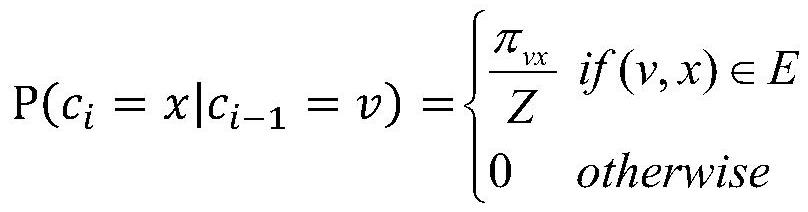Patents
Literature
39 results about "Associated Study" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
An indication that one study is related to another. (NCI)
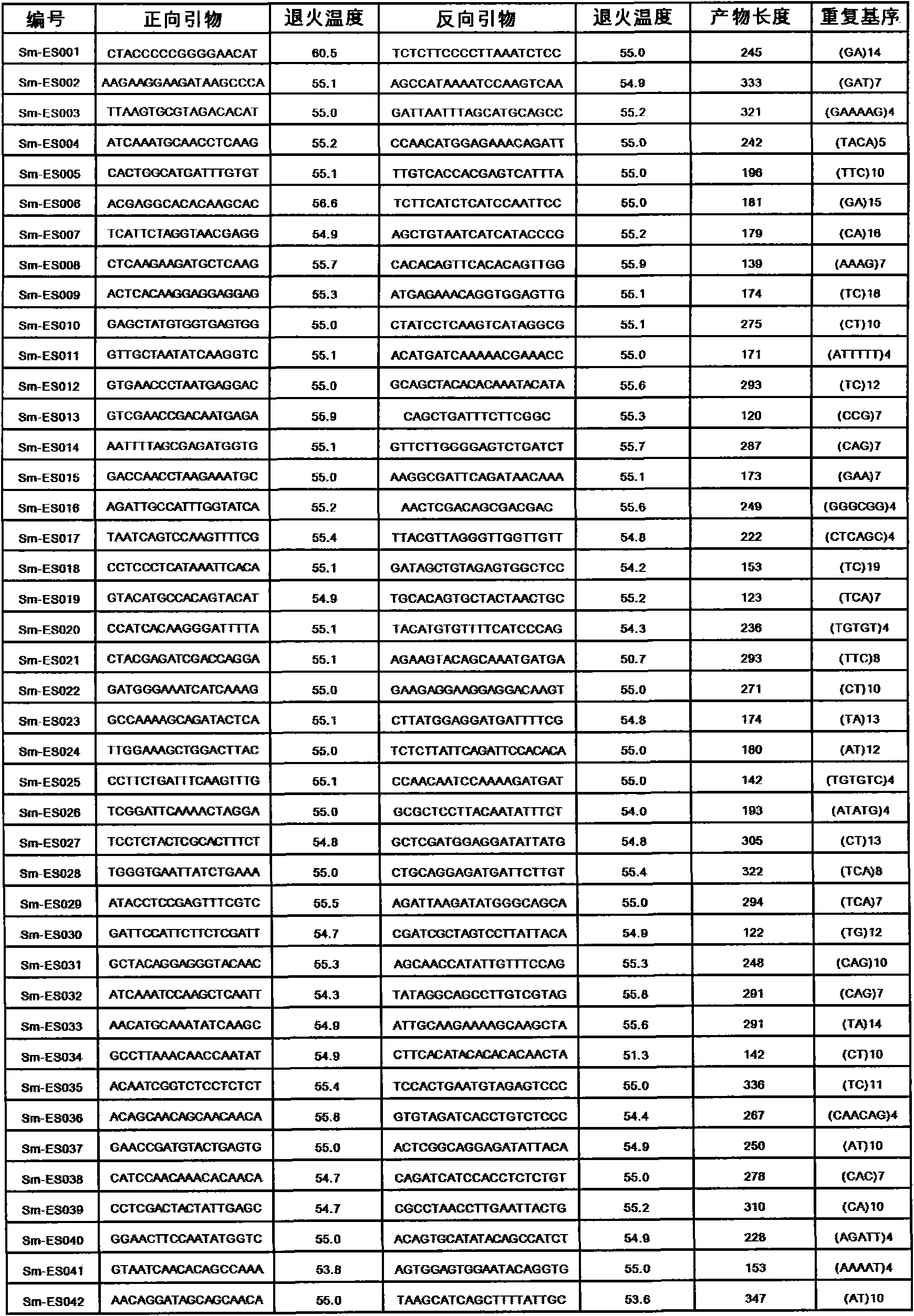
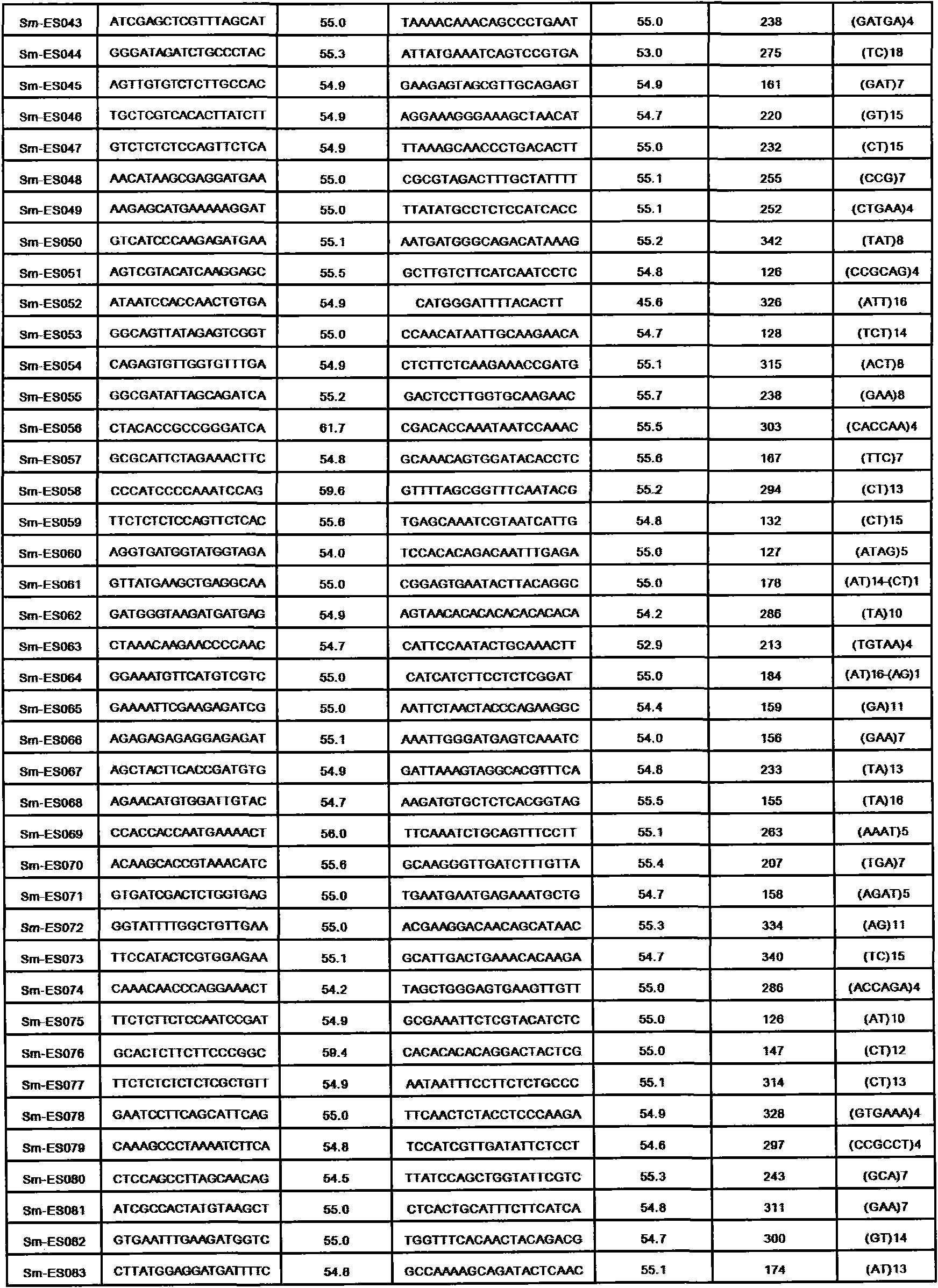
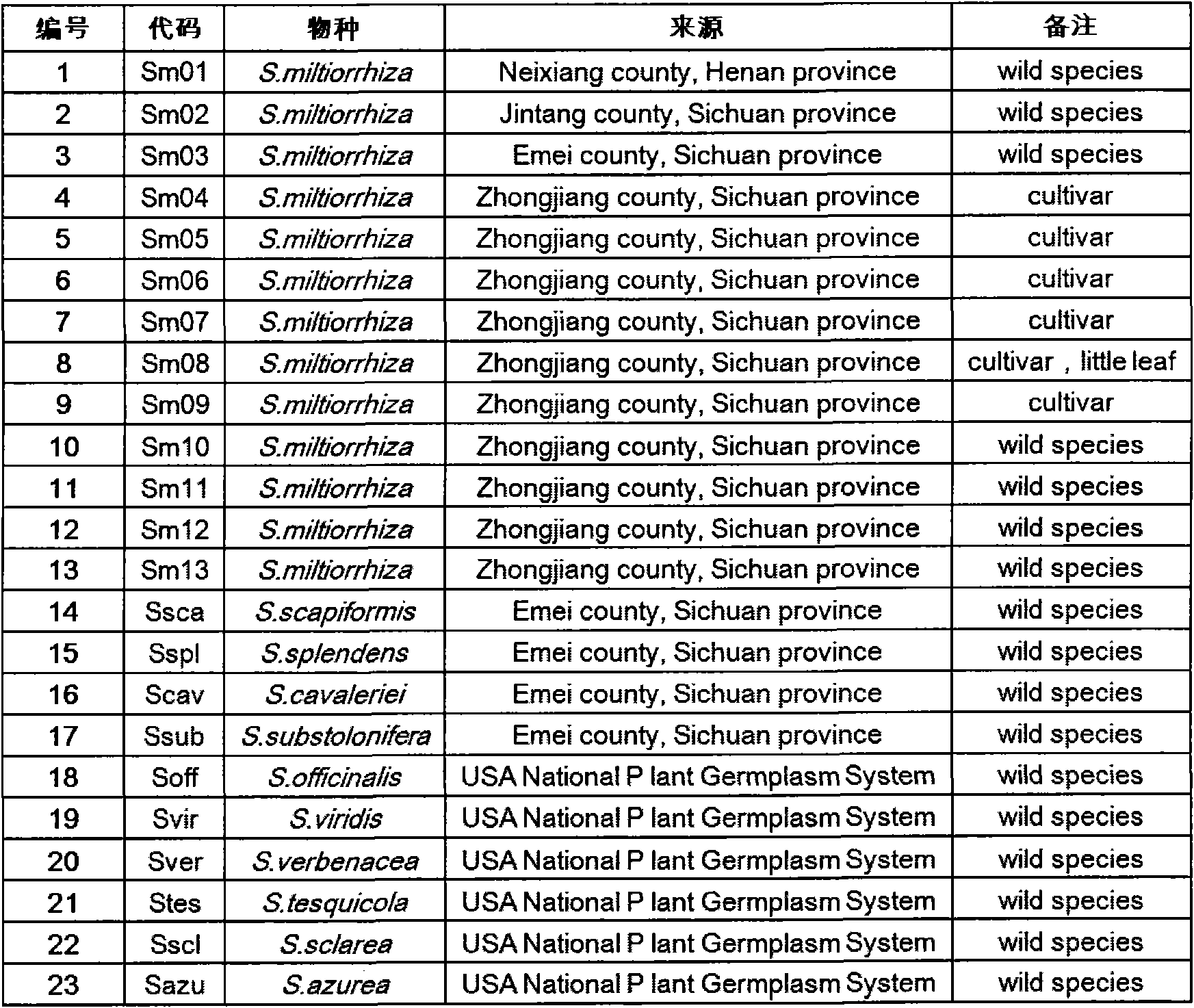
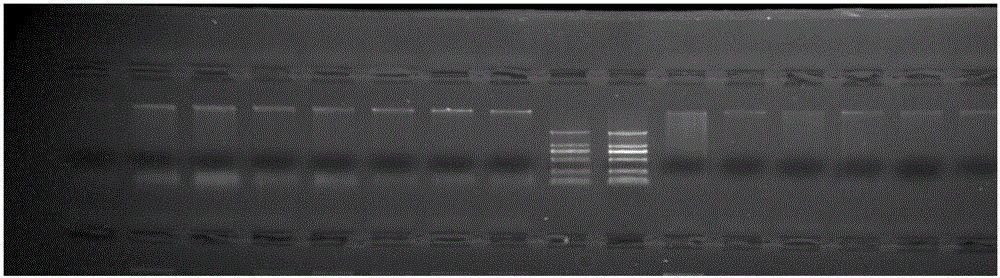
Method for preparing salvia miltiorrhiza EST-SSR molecular mark, specific primer and application thereof
The invention discloses a method for preparing a salvia miltiorrhiza EST-SSR molecular mark. The method mainly comprises the following steps of obtaining and pre-processing a salvia miltiorrhiza EST sequence, screening a SSR locus in the salvia miltiorrhiza EST sequence, designing and synthesizing the salvia miltiorrhiza EST-SSR primer, screening the salvia miltiorrhiza EST-SSR primer and the like. The method excavates the SSR locus aiming at the salvia miltiorrhiza EST and designs the PCR primer according to the flanking sequence of the SSR locus, and on the basis of this, the salvia specificEST-SSR mark system is established; and the method lays a solid foundation for genetic diversity evaluation, genetic relationship or idioplasmatic identification, genetic linkage description, molecular mark auxiliary selection, gene mapping cloning, functional genome analysis, related researches and the like of related species of the salvia miltiorrhiza and the salvia. The invention also discloses the EST-SSR locus specific primer obtained by the method for preparing the salvia miltiorrhiza EST-SSR molecular mark and the application thereof.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
Method for developing meconopsis SSR (simple sequence repeat) primers on basis of transcriptome sequences
PendingCN106834510ASolve the problem of few SSR primersAdd raw dataMicrobiological testing/measurementAgricultural scienceMeconopsis
The invention relates to a method for developing meconopsis SSR (simple sequence repeat) primers on the basis of transcriptome sequences. The method includes particular steps of (1), constructing transcriptome libraries; (2), carrying out sequencing, splicing sequenced sequences to obtain a complete transcriptome by the aid of software Trinity, utilizing each transcript with the longest genes as Unigene and carrying out bioinformatics analysis on Unigene sequences; (3), carrying out SSR detection on the Unigene by the aid of software MISA1.0; (4), designing SSR primers by the aid of software Primer3 and identifying the polymorphism of the primers. The method has the advantages that raw data for developing the primers can be greatly increased by the aid of the obtained transcriptome sequences; SSR markers can be developed on a large scale in bioinformatics ways, and accordingly the development efficiency can be greatly improved; the primers with the SSR markers are novel markers which are stably available, and can be directly used for related research on meconopsis plants, and accordingly the problem of scarceness of meconopsis SSR primers can be solved.
Owner:SOUTHWEST FORESTRY UNIVERSITY
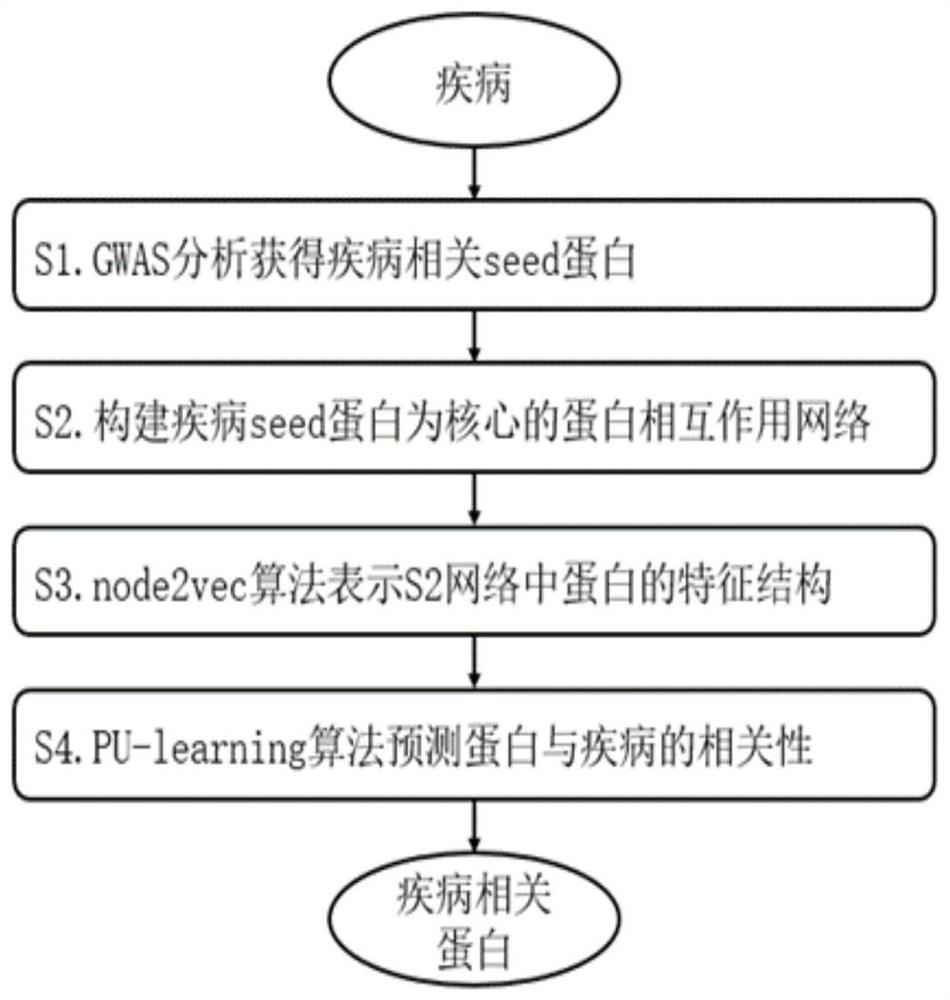
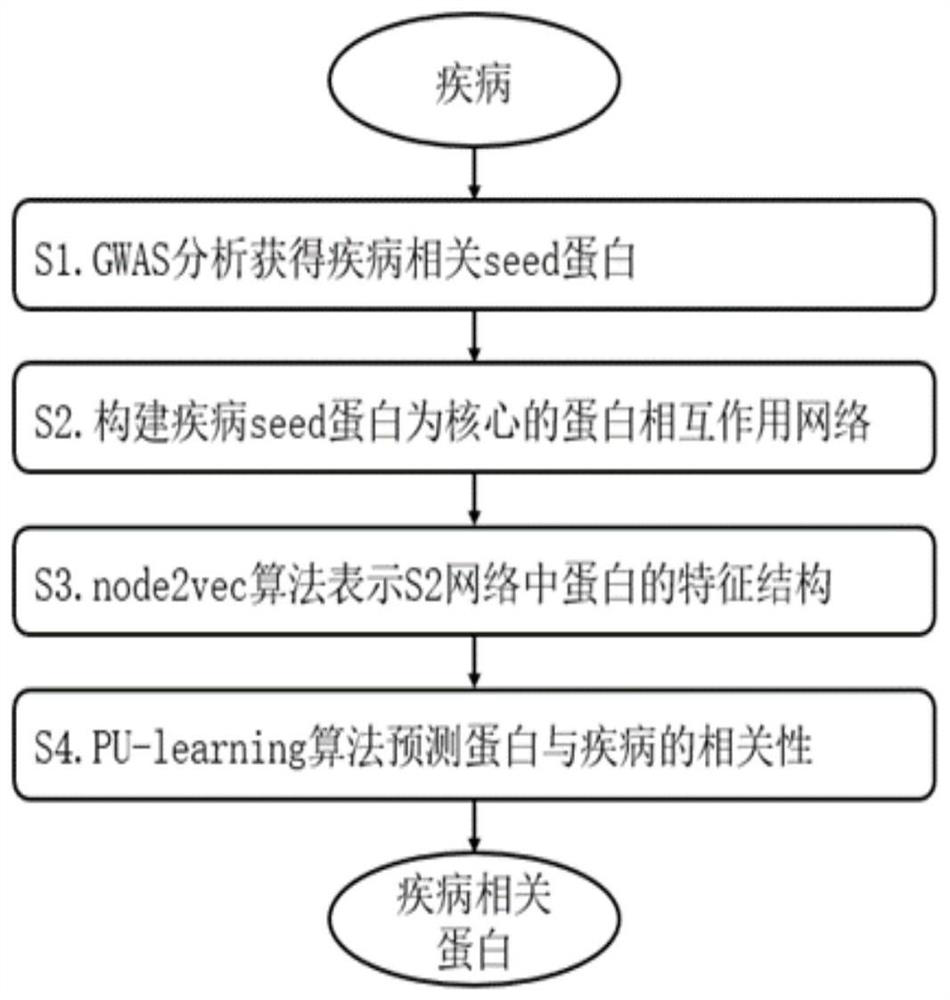
Method for screening disease-related proteins based on complex network
ActiveCN111640468AImprove accuracyReduce workloadBiostatisticsMachine learningDiseaseProtein Interaction Networks
The invention discloses a method for screening disease-related proteins based on a complex network. The method comprises the following steps: 1) acquiring a seed gene related to a target disease; 2) based on a protein-protein interaction database, constructing a protein interaction network taking the seed gene as a core; 3) extracting characteristic data of the protein in the protein interaction network; 4) taking the characteristic data of the protein as training data, and training by adopting a machine learning algorithm to obtain a PU classifier; and 5) predicting the protein related to thetarget disease in the protein interaction network according to the PU classifier. The method can quickly and efficiently identify protein related to a disease, and is helpful for biomedical experts to carry out experimental verification or related researchers to carry out work.
Owner:天士力国际基因网络药物创新中心有限公司
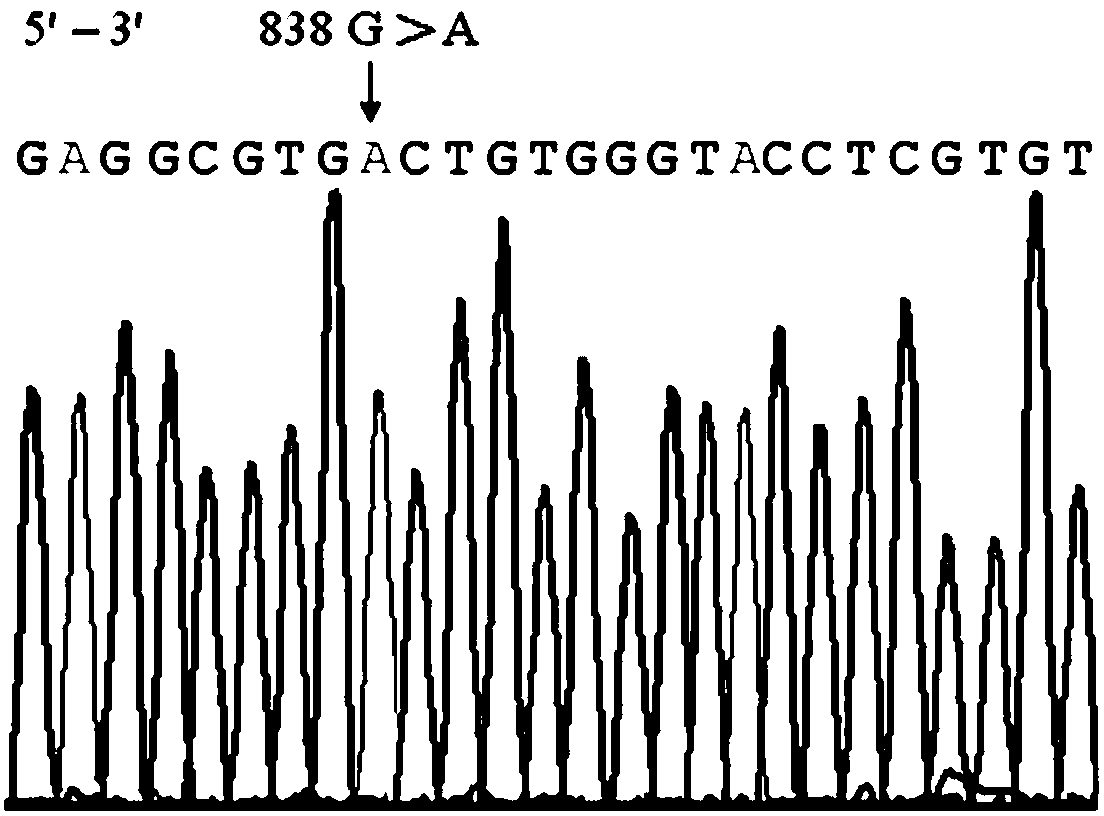
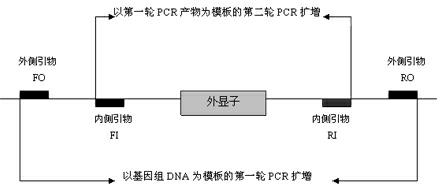
Rh blood group DEL phenotype RHD838>A allele and detection method thereof
InactiveCN103937806AExtensive scientific research application valueMicrobiological testing/measurementFermentationRhD antigenRegioselectivity
The invention relates to an RHD838>A allele and a detection method thereof, and belongs to the field of analysis and detection technologies in molecular biology. A primer sequence which can amplify a target part including a mutant gene sequence is designed by a PCR-specific primer technology, and PCR amplification selectively and specially aiming at the specific RHD838>A allele is performed on the zone of the target part including the mutant gene sequence by virtue of a gene amplification method. According to the detection method, the gene level is detected by virtue of a molecular biological method which can be used for greatly improving the sensitivity and specificity of the detection and is simple, convenient and rapid. Since different nationalities have difference in the RHD gene, the detection method is designed specially aiming at the RHD838>A allele according to the RHD genetic molecule background of Chinese on the basis of relative researches. The RHD838>A allele and the detection method thereof have significant clinical meaning in overcoming the shortage of the serological technique, and also have wide scientific research and application value.
Owner:WUXI NO 5 PEOPLES HOSPITAL
GJB2 gene p.V37I mutation diagnostic kit for diagnosing late-onset deafness
InactiveCN102304584AEstablish priority areas of applicationMicrobiological testing/measurementGenes mutationFull Term Neonate
The invention relates to application of GJB2 gene p.V37I mutation to the preparation of products for diagnosing late-onset deafness. The GJB2 gene p.V37I mutation is p.V37I exclusive mutation. The invention also provides a GJB2 gene p.V37I mutation diagnostic kit for diagnosing the late-onset deafness and application of the kit. The GJB2 gene p.V37I mutation diagnostic kit has the advantages that: a common molecular identifier, namely the GJB2 gene p.V37I exclusive mutation related to the late-onset deafness highly is discovered at home and abroad for the first time; a molecular identifier-based gene screening method and the preferential application range of the molecular identifier in the screening of neonates are determined; and the alarm time of the late-onset deafness caused by the GJB2 gene p.V37I exclusive mutation is advanced to a neonate period, and the GJB2 gene p.V37I mutation diagnostic kit is widely applied to the early detection, intervention and rehabilitation of genetic susceptible late-onset deafness, the screening and diagnosis of deafness genes, late-onset deafness hearing-gene combined screening and other relevant study fields.
Owner:XIN HUA HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
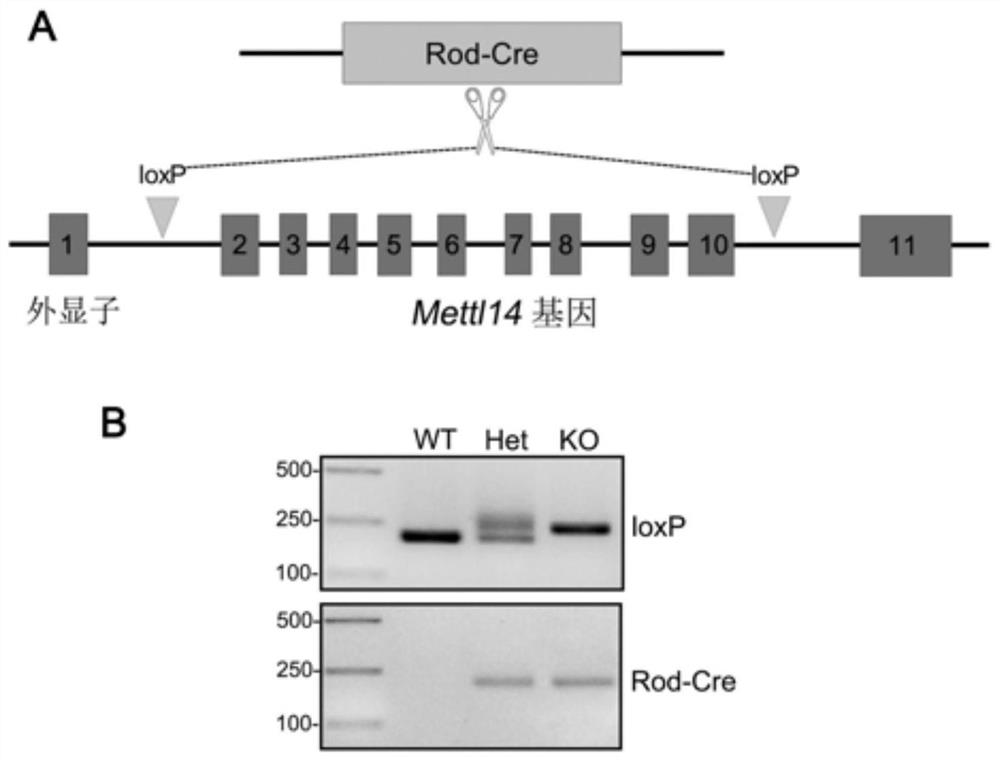
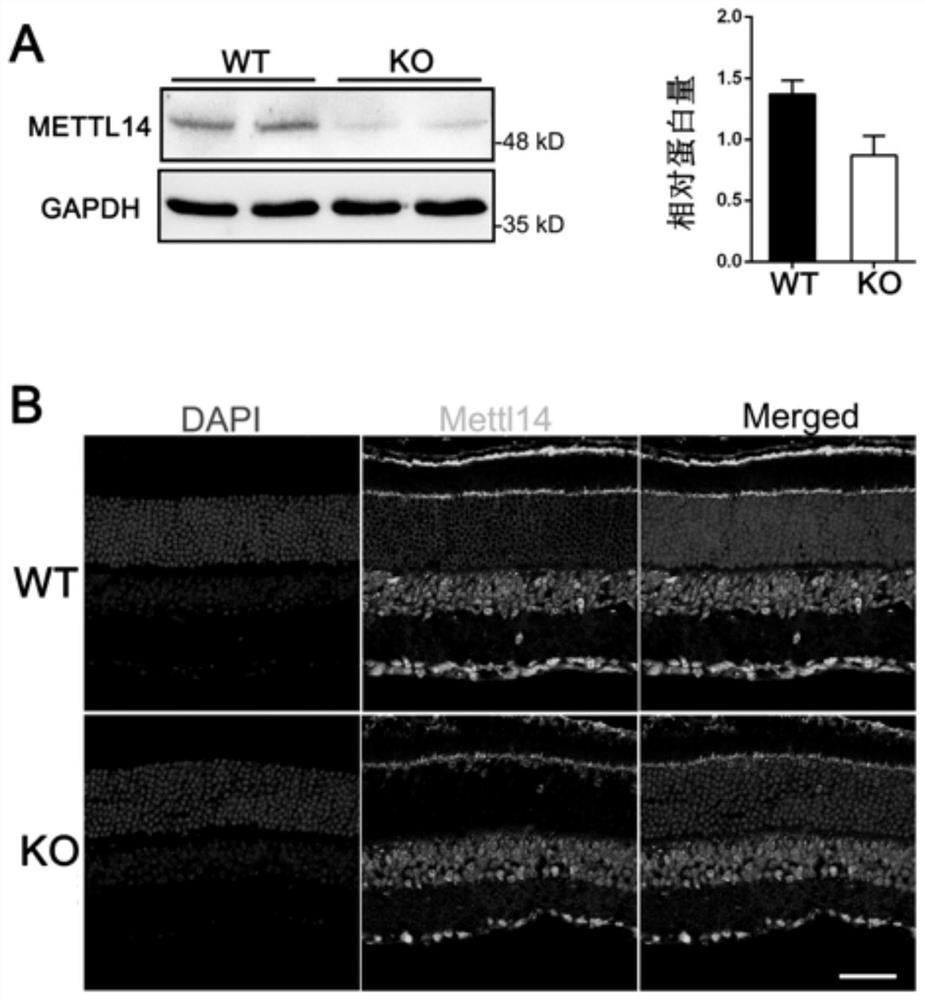
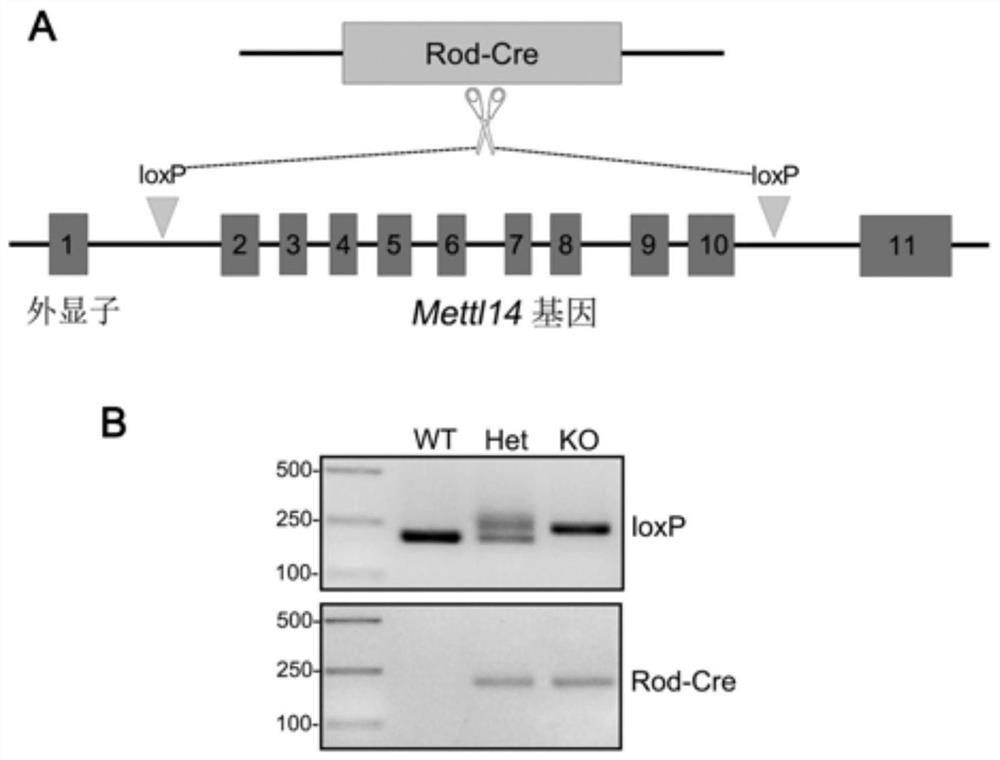
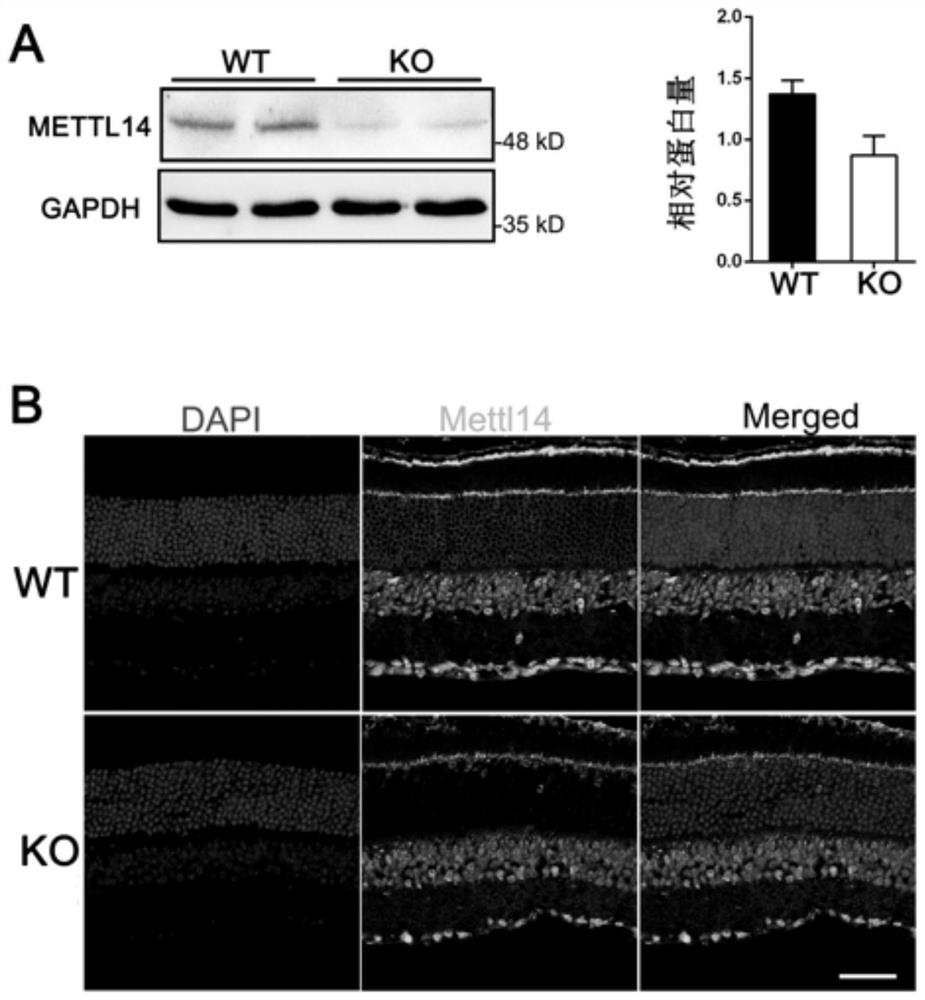
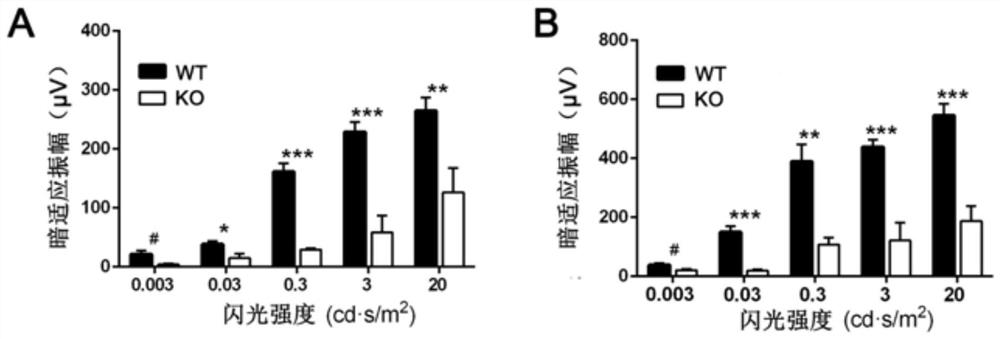
Method for constructing retinitis pigmentosa disease model, application and breeding method
The invention discloses a method for constructing a retinitis pigmentosa disease model, application and a breeding method, and relates to the technical field of gene editing. According to the method, a Mettl14 gene of a target animal is modified, so that the target animal shows typical retinitis pigmentosa disease characteristics, and the target animal can be used as the retinitis pigmentosa disease model. According to the method, a new disease model with retinitis pigmentosa disease characteristics can be constructed. The disease model can be used for related research on the pathogenesis process and mechanism of the retinitis pigmentosa disease, can also be used for screening related drugs of the retinitis pigmentosa disease, and has a wide application prospect.
Owner:SICHUAN PROVINCIAL PEOPLES HOSPITAL
Cervinae activin A protein and preparation method and application thereof
ActiveCN109293764AImprove researchGood application prospectHormone peptidesNervous disorderBiotechnologyGene
The invention relates to the field of molecular biology and particularly relates to a cervinae activin A protein and a preparation method and application thereof. The activin A protein is composed oftwo cervinae activin beta A subunits isolated from cervinae animals. The study on the activin A protein is still relatively rare in the cervinae. The preparation method acquires the isolated gene sequence, improves its high-purity protein expression method, provides research basis and novel ideas for related research and greatly increases the research and application prospect of the activin A protein.
Owner:INST OF SPECIAL ANIMAL & PLANT SCI OF CAAS
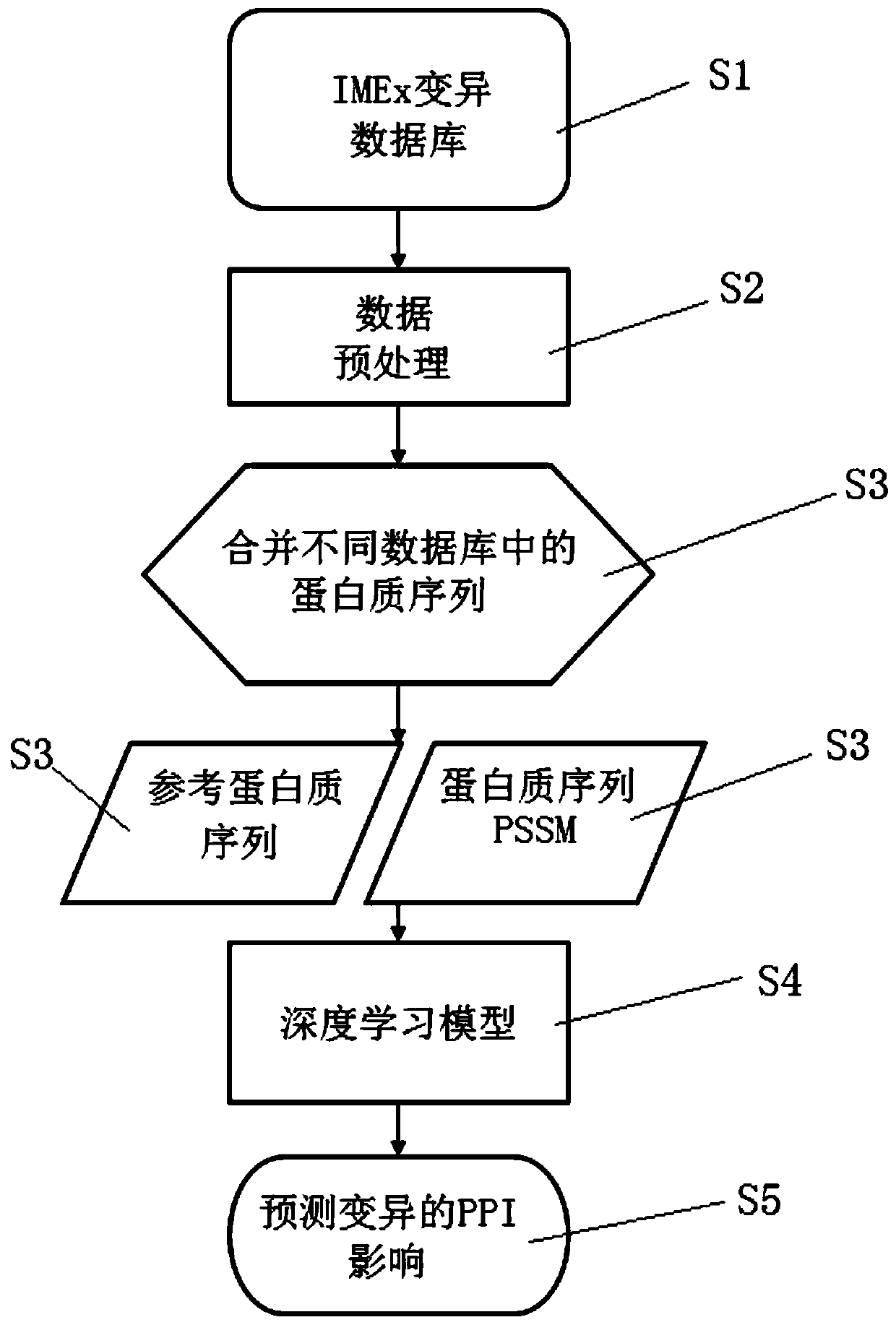
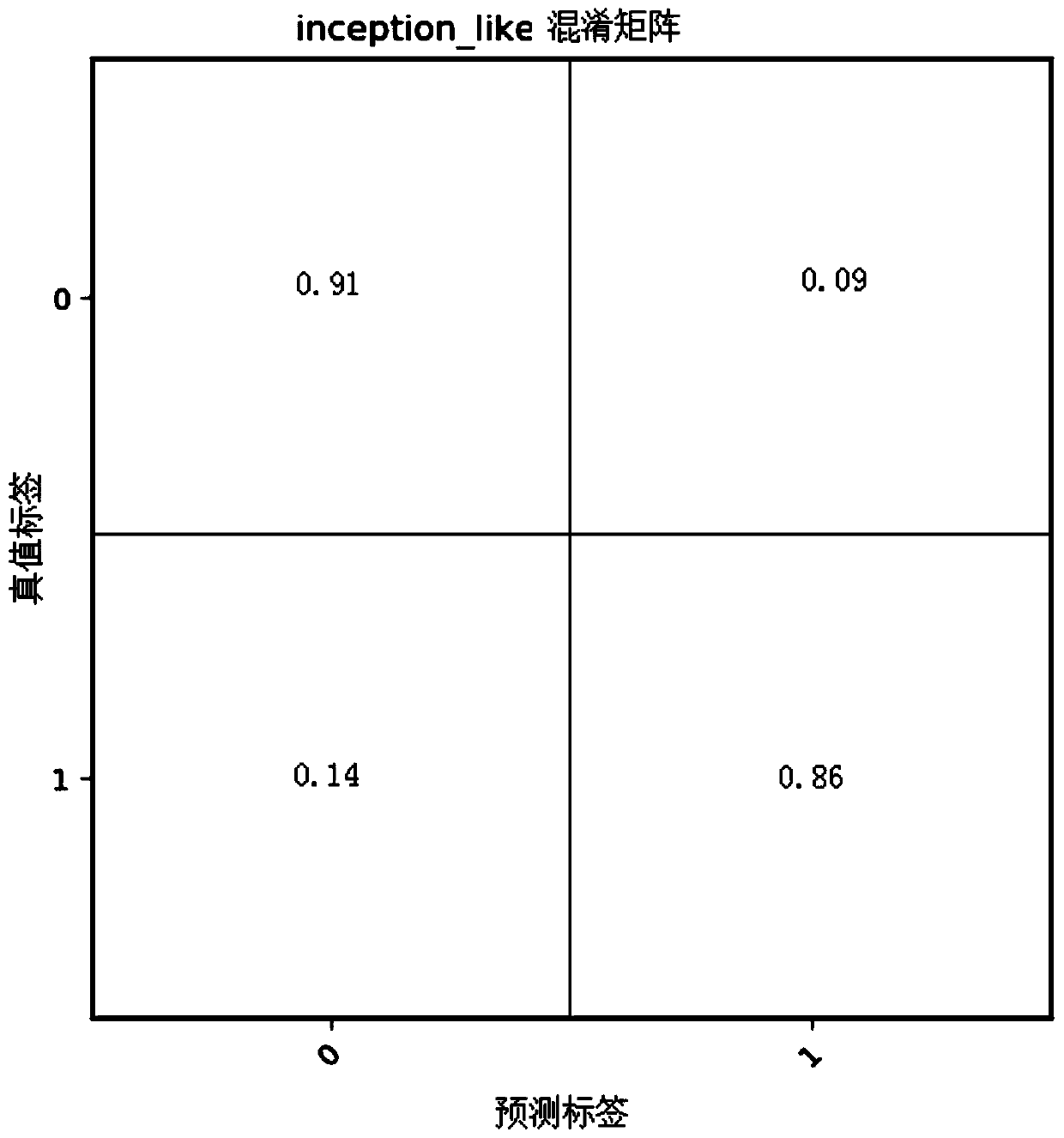
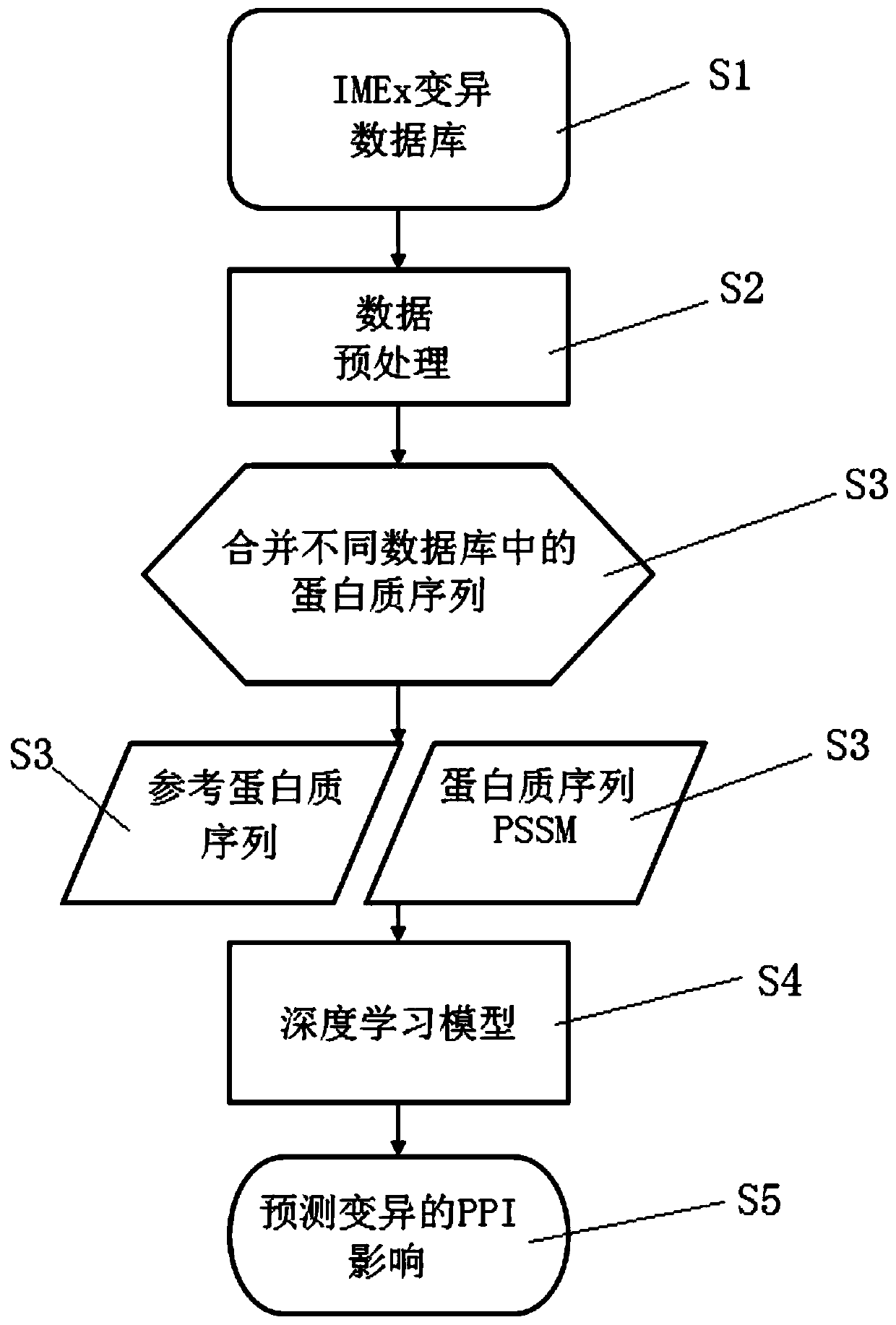
Method for judging influence on protein interaction based on mutation information
The invention discloses a method for judging the influence on protein interaction based on mutation information. The method is a judgment tool (MIPPI) for judging whether single-point mutation in a protein can generate negative influence on the protein interaction of the original gene, and comprises three parts of data collection and screening, feature selection and extraction, and model establishment. According to the technical scheme, an intuitive auxiliary judgment standard for the influence of mutation on protein interaction can be provided for related researchers of gene and protein mutation, the influence on protein interaction caused by mutation is judged mainly based on protein sequence mutation information, and judgment on the severity degree of protein mutation can be improved.
Owner:SHANGHAI MENTAL HEALTH CENT (SHANGHAI PSYCHOLOGICAL COUNSELLING TRAINING CENT)
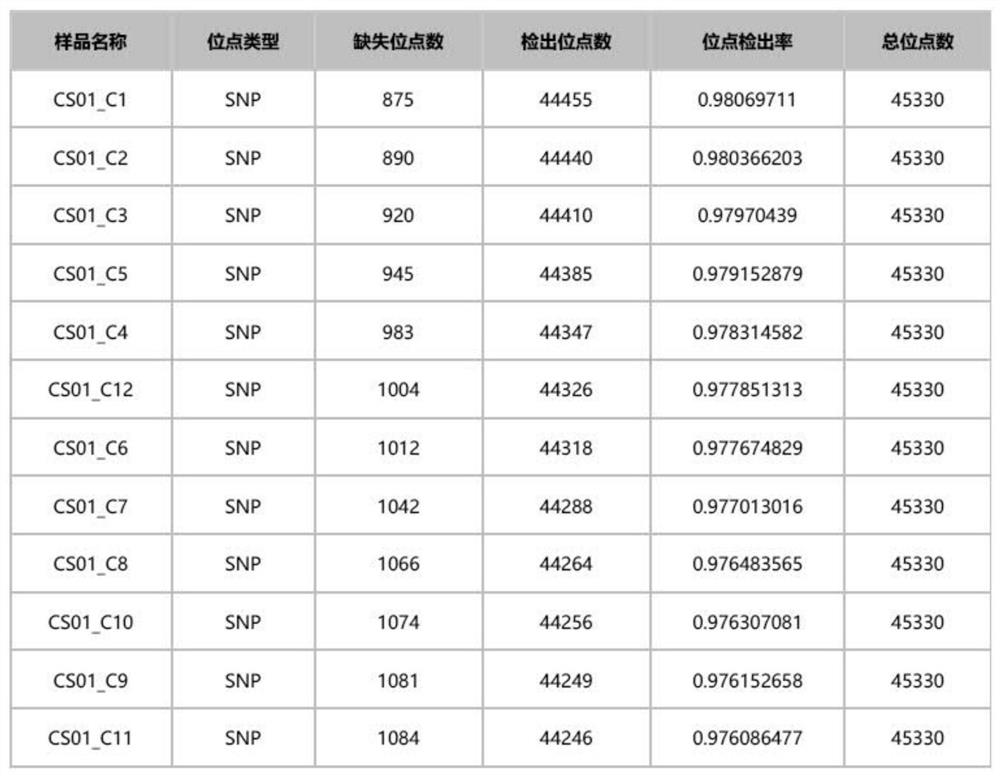
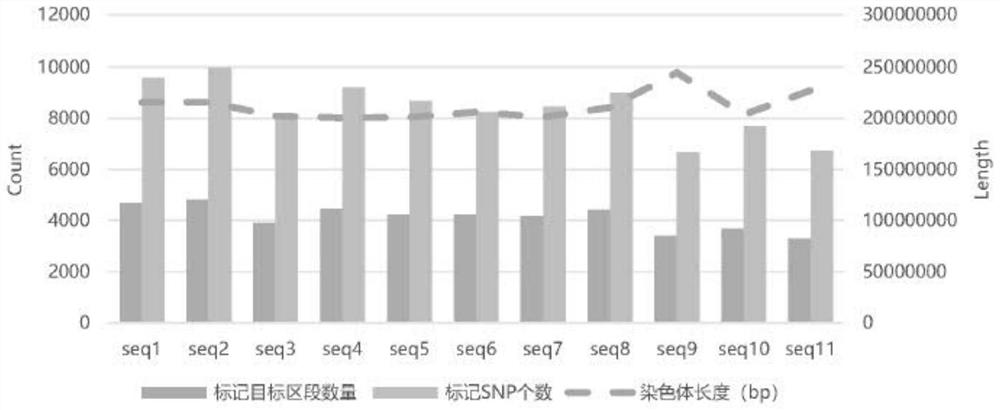
Sika deer whole genome SNP molecular marker combination, SNP chip and application
ActiveCN114292924AImprove accuracyRich in variationFood processingMicrobiological testing/measurementBiotechnologyGenetics
The invention discloses a sika deer whole genome SNP molecular marker combination, an SNP chip and application, and relates to the technical field of molecular marker development. Comprising 43316 SNP molecular markers. According to the SNP molecular marker combination, when the sika deer sample data volume is 1.5 Gb, the site detection rate is 95% or above, and it is indicated that the SNP molecular marker combination is large in useful information amount and has high utilization value. In addition, the SNP molecular marker combination developed by the invention is uniformly distributed on 33 chromosomes of a sika deer whole genome, so that the accuracy of related research application can be improved.
Owner:INST OF SPECIAL ANIMAL & PLANT SCI OF CAAS
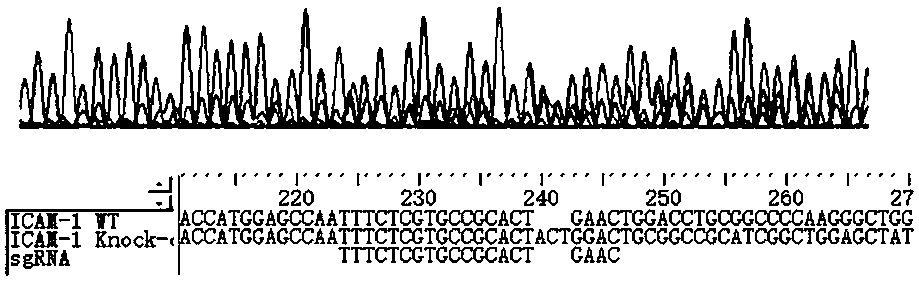
ICAM-1 (Intercellular Adhesion Molecule 1) gene knockout tumor cell strain and application thereof
InactiveCN108467864AImprove biological activityEfficient targetingGenetically modified cellsNucleic acid vectorHuman DNA sequencingCytokine
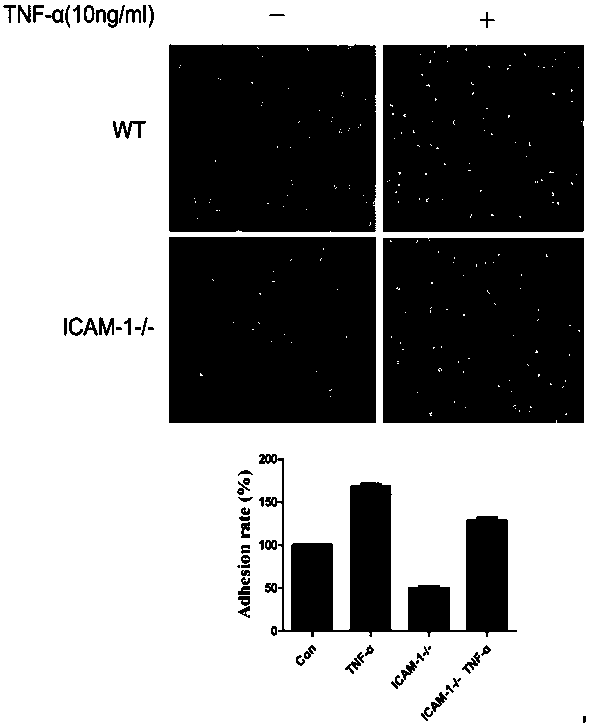
The invention relates to related study fields of cytobiology, molecular biology and the like and discloses an ICAM-1 (Intercellular Adhesion Molecule 1) gene knockout tumor cell strain and applicationthereof. The invention discloses sgRNA (Small Guide Ribonucleic Acid) which targets to a PAM (Protospacer Adjacent Motif) site of an ICAM-1 gene of a human genome; through transfection recombinant CRISPR plasmid transfection, the ICAM-1 gene of a HelA cell can be efficiently knocked out; through resistance screening with puromycin, a conventional limiting dilution method is not used in the screening process, but a relatively convenient and concise monoclonal picking method is used instead, and thus the experiment cycle is greatly shortened. With the ICAM-1 knockout cell strain, a novel research platform is established for studying adhesion and metastasis process of tumor cells, experiments show that the adhesion property of HeLa is remarkably degraded after ICAM-1 is knocked out, molecular mechanisms of mutual actions of ICAM-1 with different cell factors can be studied, and meanwhile treatment mechanisms and molecular targets related to anti-metastasis medicines can be well studied.
Owner:FUZHOU UNIV
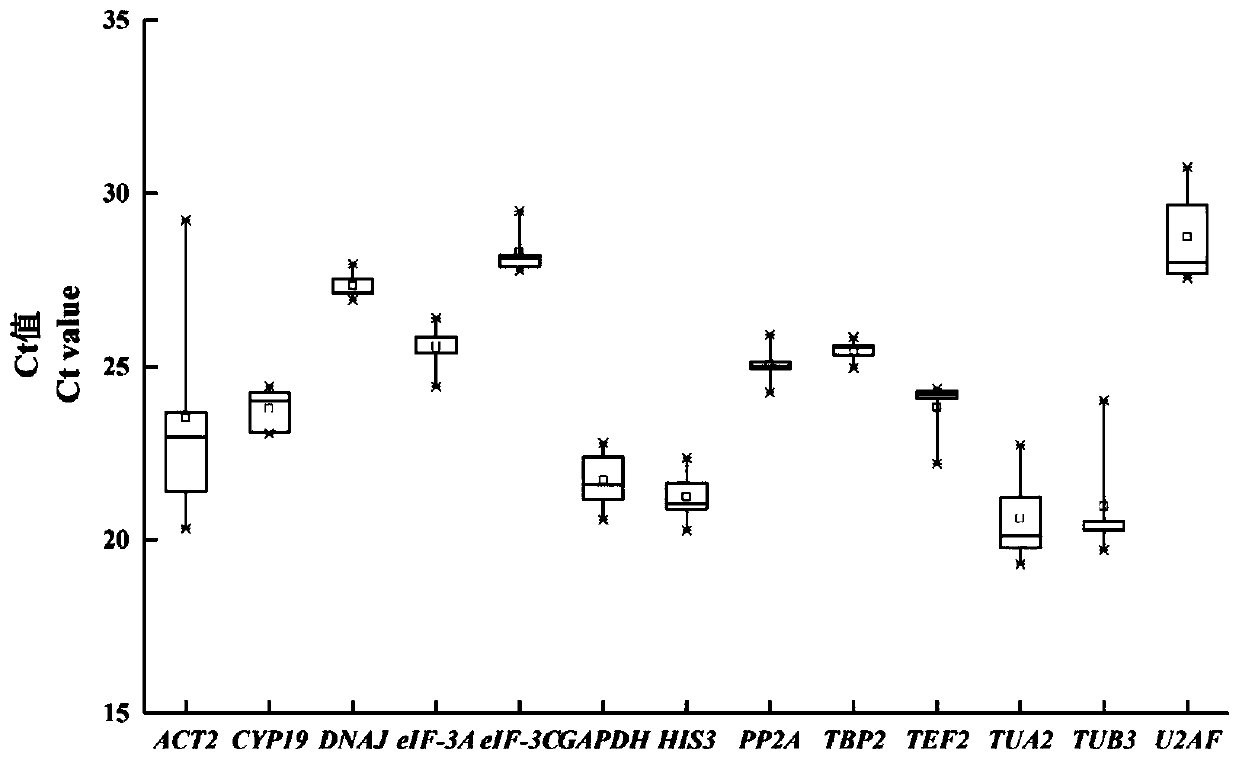
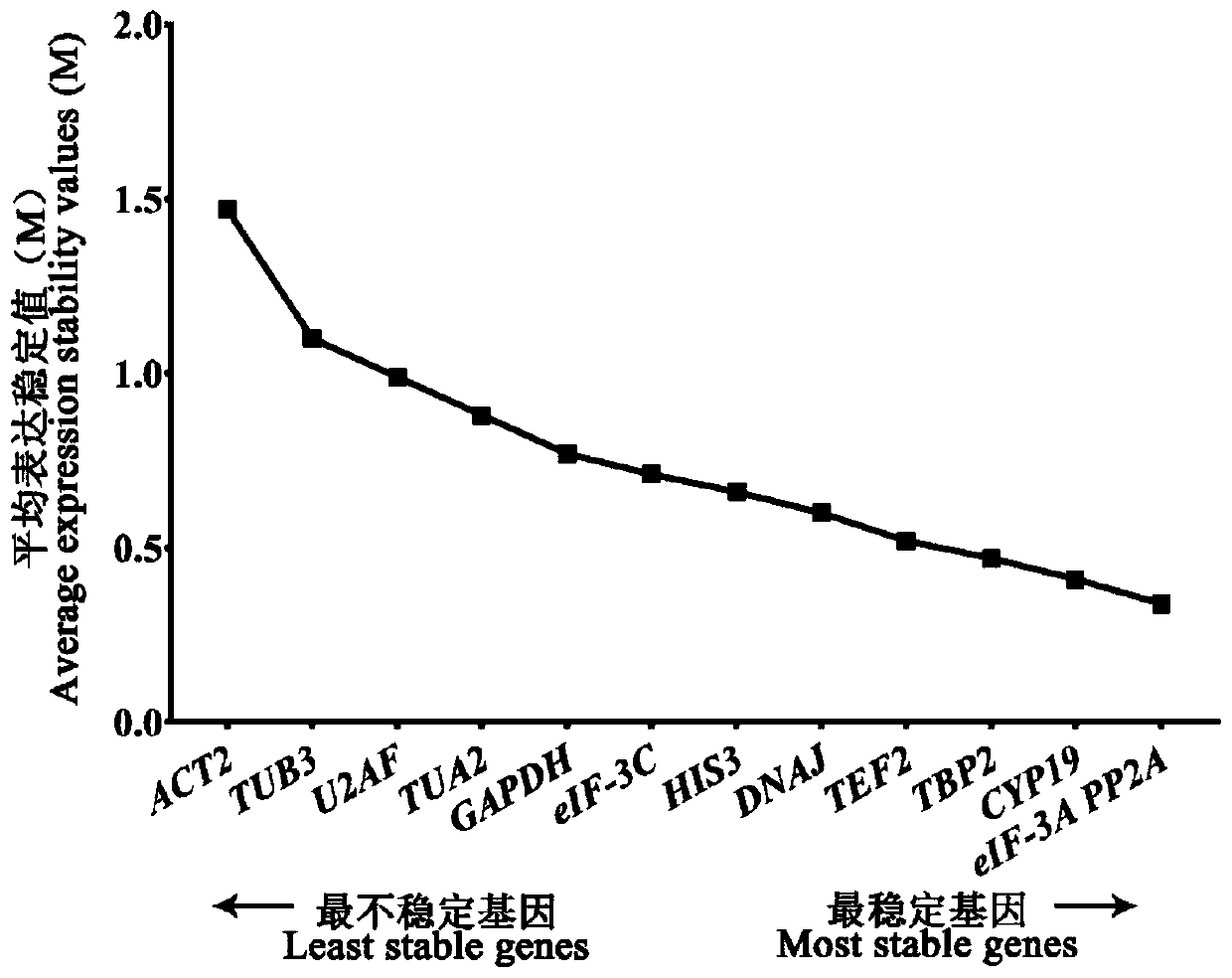
Fluorescent quantitative reference genes of different tissues of siberian wildrye and primers and application thereof
InactiveCN110283933AIncrease credibilityA large amountMicrobiological testing/measurementDNA/RNA fragmentationReference genesNucleotide
The invention provides fluorescent quantitative reference genes of different tissues of siberian wildrye: TBP2 and PP2A, the nucleotide sequences of which are as shown in sequence tables SEQ ID NO.1 and SEQ ID NO.2. The two reference gene sequences are originated from siberian wildrye transcriptome sequences and the reference genes are higher in specificity and stability compared with those of universal reference genes on other species. Meanwhile, the two reference genes screened are the most stable reference genes of siberian wildrye in different tissues. Data is true and reliable, thereby laying a foundation for deeply digging the functional genes of siberian wildrye in later period. The fluorescent quantitative reference genes not only solve the current situation that no reference genes are available in current siberian wildrye gene expression analysis, but also can improve the reliability of detection efficiency and detection result. The two reference genes are newly developed genes, so that the quantity of reference genes of plants is enriched, and reference can be also provided for associated researches on other wheat families and lyme grass plants.
Owner:LANZHOU UNIVERSITY
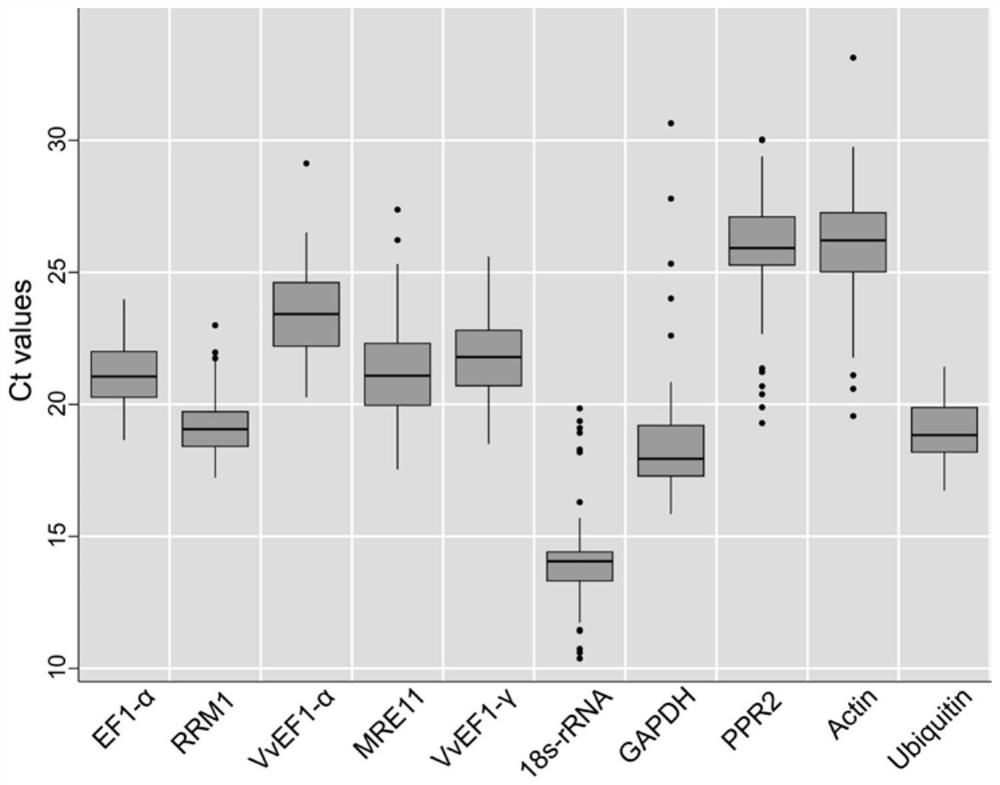
QRT-PCR reference gene of grapes as well as primers and application of qRT-PCR reference gene
ActiveCN112011643AMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyReference genes
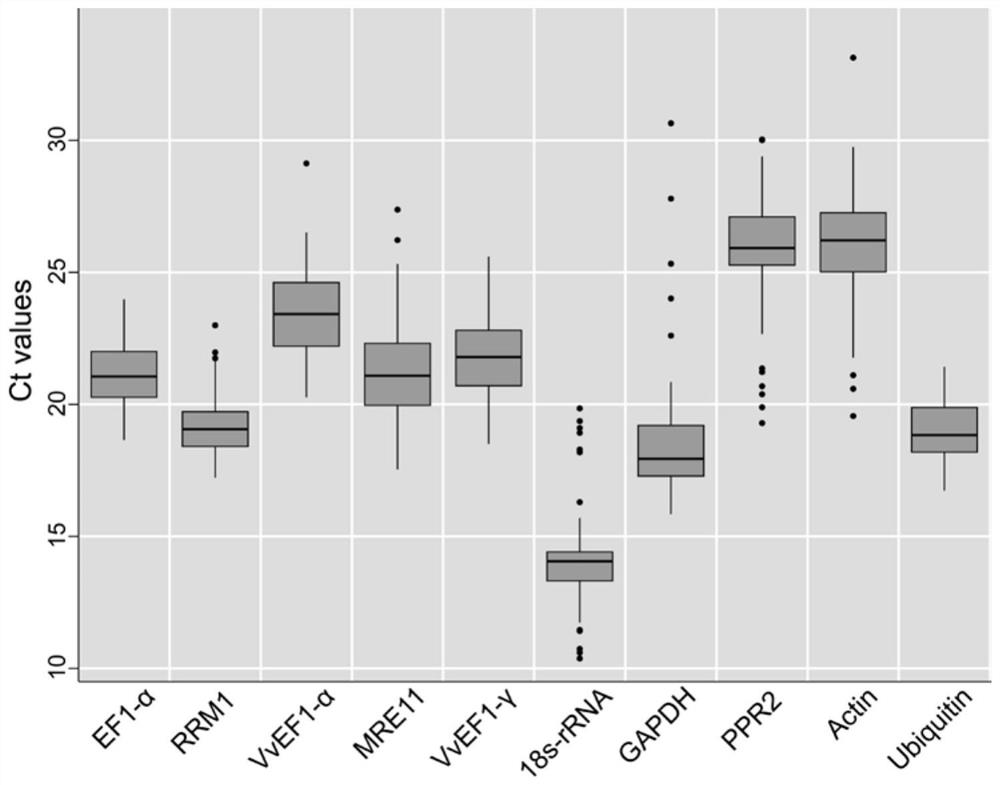
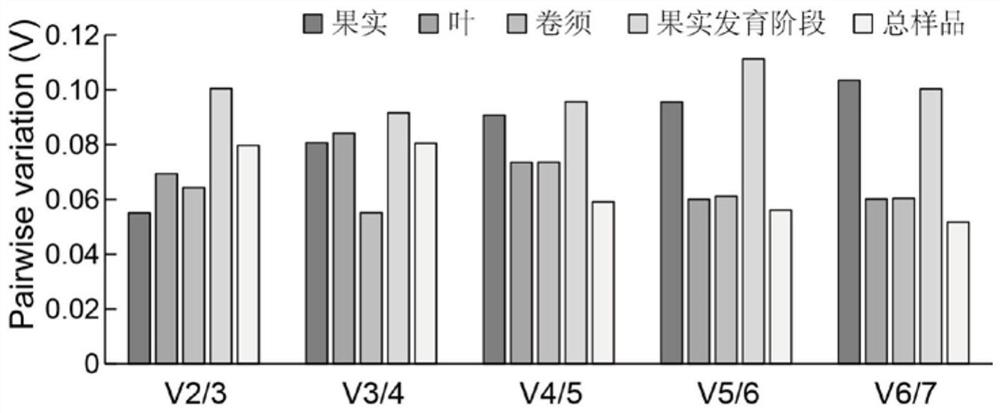
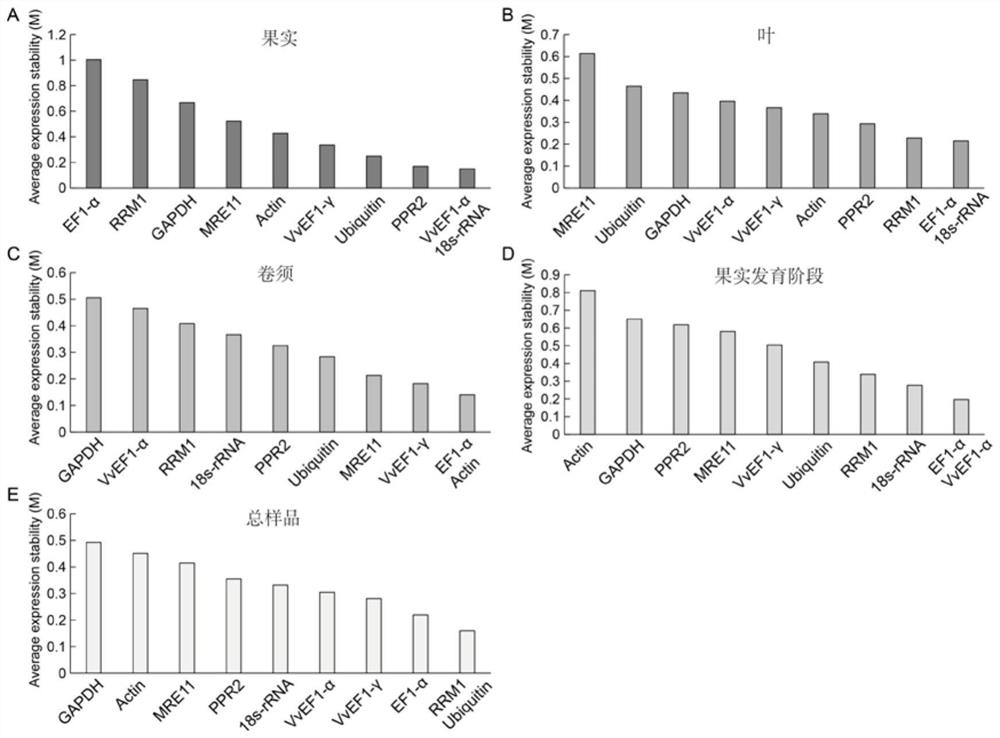
The invention discloses a qRT-PCR reference gene of grapes as well as primers and application of the qRT-PCR reference gene. A new grape reference gene RRM1 is identified for the first time, and qRT-PCR primers aiming at the gene are designed. According to transcriptome data in an earlier stage, the stability of seven candidate reference genes, namely, Actin, 18s-rRNA, GAPDH, VvEF1-gamma, VvEF1-alpha, EF1-alpha and Ubiquitin, and three newly explored candidate reference genes, namely, RRM1, PPR2 and MRE11 in fruits, leaves and beard curls of seven different grape varieties and Kyoho fruit samples of nine different development stages is analyzed. Results show that the number of optimal reference genes for the grape sample is two, the selection of the optimal reference genes for different samples is different, the optimal reference genes in grape leaf tissues are RRM1 and EF1-alpha, and the optimal candidate reference genes are RRM1 and Ubiquitin when all the samples are contained. The qRT-PCR kit can provide important reference for related researchers to carry out qRT-PCR work in grapes.
Owner:HENAN UNIV OF SCI & TECH
Method for detecting adsorption completeness of recombinant novel coronavirus vaccine
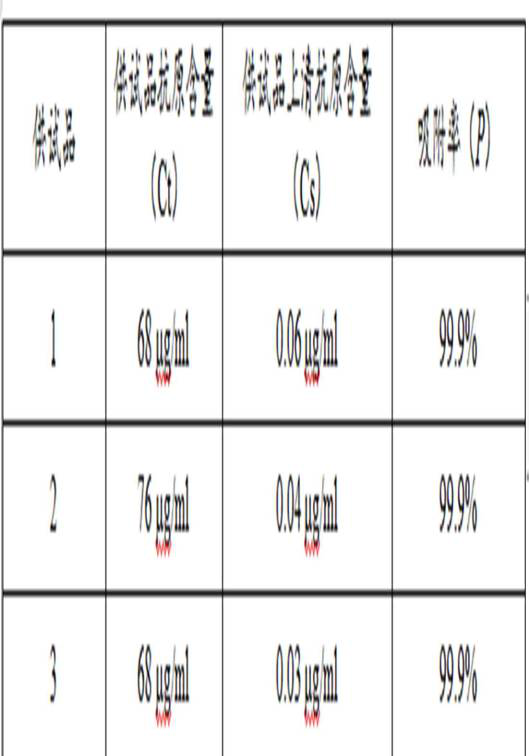
ActiveCN111735967AExperimental results are reliableBiological material analysisBiological testingCoronavirus vaccinationTest sample
The invention discloses a method for detecting the adsorption completeness of a recombinant novel coronavirus vaccine, and belongs to the technical field of biology. The method comprises the followingsteps: pre-coating a detection plate with an RBD antibody to obtain a coated plate; closing the coating plate, and incubating to obtain a closed elisa plate; carrying out gradient dilution on a reference substance; desorbing a test sample, and preparing a desorbed diluent and a non-desorbed supernatant; respectively adding the reference substance, the test sample desorption diluent and the test sample non-desorbed supernatant with various concentrations into plate holes, and incubating by taking the sample diluent as a negative control; adding an enzyme labeled secondary antibody, and incubating; adding a substrate color developing solution, and developing in a dark place; and terminating color development, reading, and counting and reading an absorbance value. The method can be used foradsorption completeness detection and related research work of the recombinant novel coronavirus vaccine semi-finished product, so that the experimental result is accurate and reliable.
Owner:天津中逸安健生物科技有限公司 +1
A PCR detection primer and kit for Riemerella anatipestifer virulent phage
ActiveCN112458202BRapid identificationMicrobiological testing/measurementMicroorganism based processesElectrophoresesRapid identification
The invention belongs to the technical field of biotechnology and diagnostic detection, and discloses a PCR detection primer for Riemerella anatipestifer virulent phage and a kit thereof, wherein the primers are shown in SEQ ID NO: 5 and SEQ ID NO: 6, The Riemerella anatipestifer virulent phage can be quickly identified simply by using a pair of specific primers to amplify the sample to be tested by PCR and then electrophoresis. At present, there is no report on the identification of Riemerella anatipestifer virulent phage based on molecular biology methods. This study can fill the gap in related research fields.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
The internal reference gene developed based on Miscanthus transcriptome sequence and its application
ActiveCN109609685BImprove stabilityImprove the correction effectMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyReference genes
The invention discloses a reference gene developed based on a transcriptome sequence of miscanthus sinensis and application thereof. The three reference gene sequences of the invention come from the transcriptome sequence of miscanthus sinensis, and have the advantages of good specificity and high stability compared with the previous reference genes commonly used in other species. At the same time, the expression analysis of SOD and CAT genes under drought stress of miscanthus sinensis proves that the developed three reference genes have better calibration capability than other general reference genes, and provides an effective reference gene calibration tool for the analysis, screening and validation of the gene expression of miscanthus sinensis in the future. In addition, the newly developed reference gene enriches the available number of plant reference genes, as well as is applied to the related research on other andropogoneae and miscanthus plants. The reference gene provided by the invention is suitable for popularization and application in the field of reference genes.
Owner:SICHUAN AGRI UNIV
Method, application and breeding method for constructing retinitis pigmentosa disease model
ActiveCN112715484BVector-based foreign material introductionAnimal husbandryOphthalmologyGene Modification
The invention discloses a method for constructing a retinitis pigmentosa disease model, an application and a breeding method, and relates to the technical field of gene editing. The method disclosed in the present invention modifies the Mettl14 gene of the target animal so that the target animal exhibits typical characteristics of the retinitis pigmentosa disease, and can be used as a model of the retinitis pigmentosa disease. The method disclosed in the invention can construct a new disease model with the characteristics of retinitis pigmentosa disease. The disease model can be used for related studies on the pathogenesis and mechanism of retinitis pigmentosa disease, and can also be used for drug screening related to retinitis pigmentosa disease, and has broad application prospects.
Owner:SICHUAN PROVINCIAL PEOPLES HOSPITAL
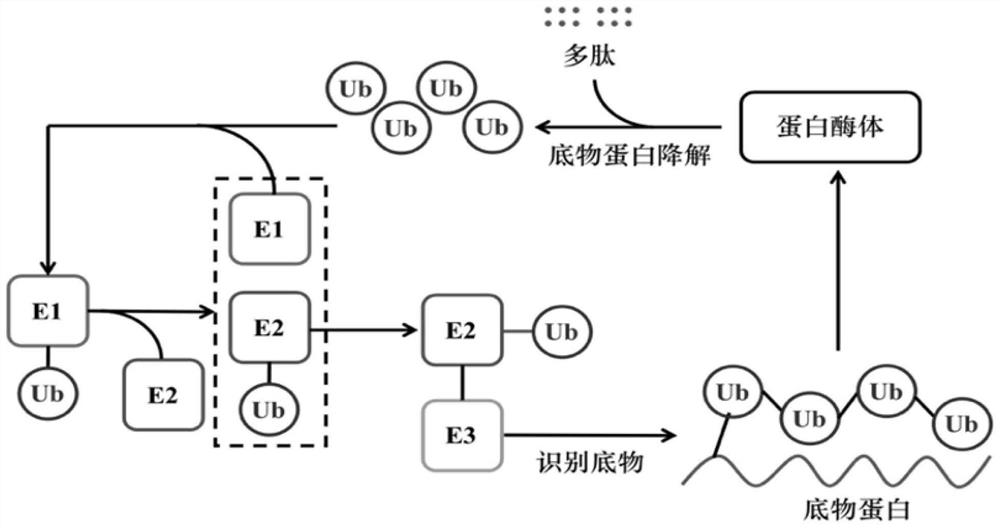
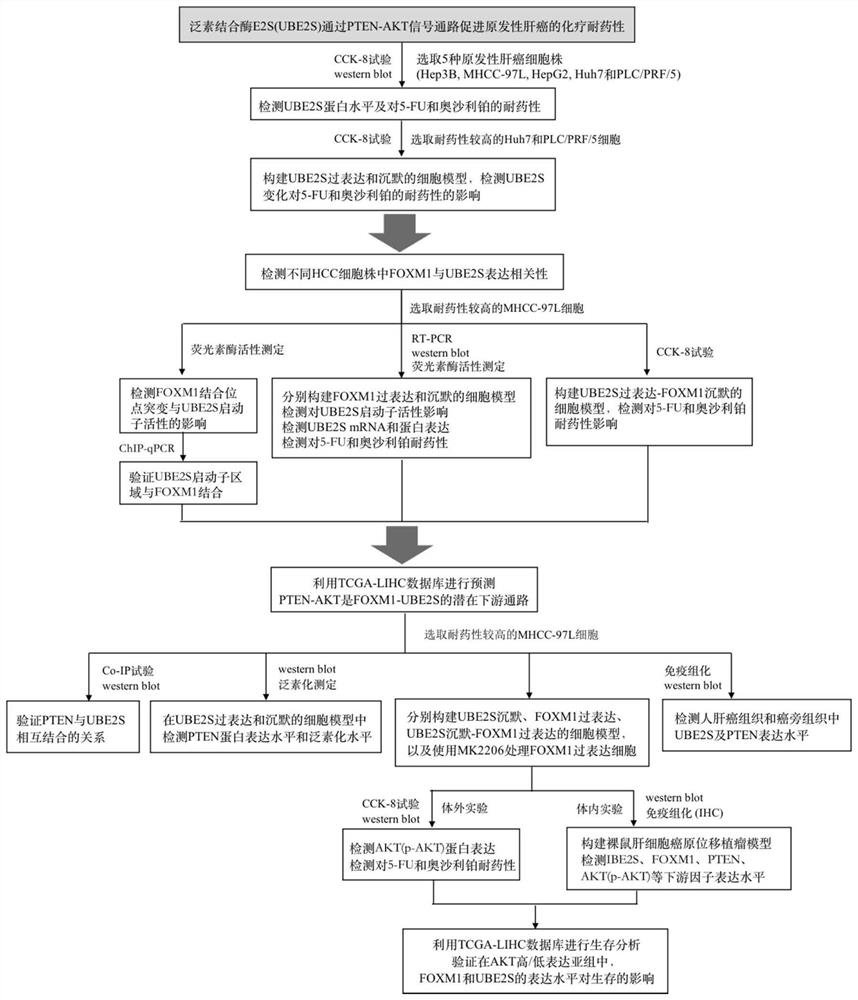
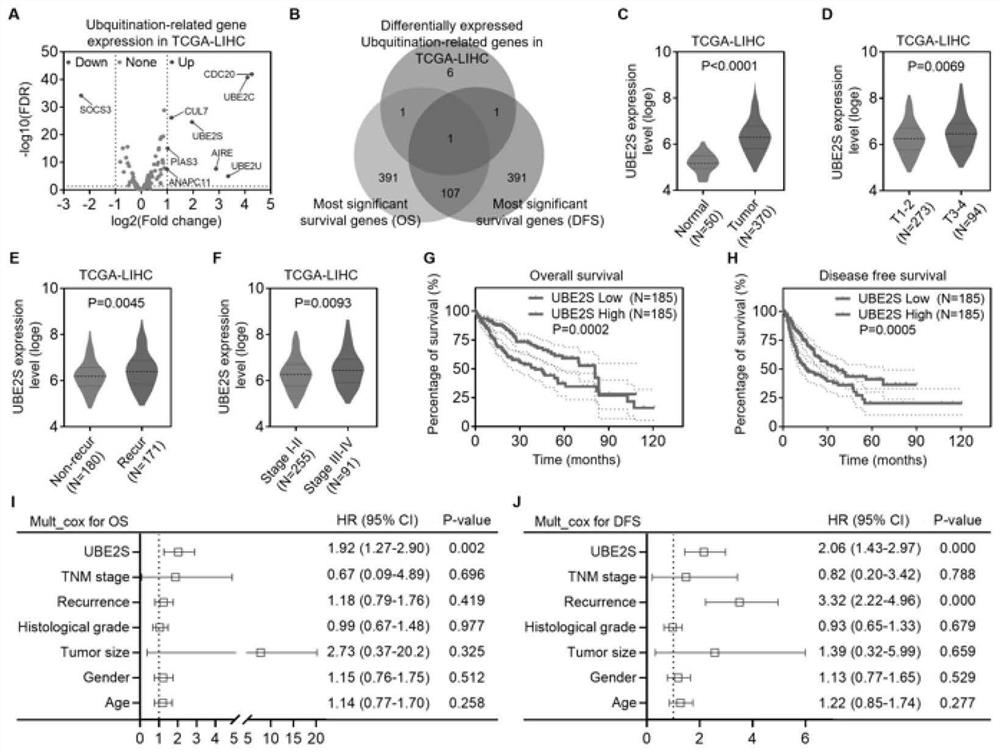
Method for verifying reason for promoting HCC chemotherapy drug resistance by regulating PTEN-AKT signal axis through UBE2S
PendingCN114561468AReduced expression levelImproved prognosisMicrobiological testing/measurementBiological testingAkt signallingOncology
The invention provides a method for verifying reasons for promoting HCC chemotherapy drug resistance by using UBE2S to regulate a PTEN-AKT signal axis. The method comprises the following processes: verifying a relationship between UBE2S in HCC and a drug-resistant pathway, silencing UBE2S in HCC cells, and verifying UBE2S and the drug-resistant pathway by using an RNA-seq technology and gene enrichment analysis; the drug resistance of UBE2S expression in the HCC cells to fluorouracil and oxaliplatin is verified; the correlation between the transcription factor FOXM1 and UBE2S expression is verified; fOXM1-UBE2S downstream molecules and the regulation and control effect thereof, in-vivo experiments and in-vitro experiments are verified. The invention verifies that abnormal high expression of UBE2S exists in chemotherapy-resistant HCC cells, and influences on chemotherapy resistance by promoting PTEN ubiquitination, reducing the PTEN protein expression level and regulating and controlling a PI3K-AKT signal axis are achieved. UBE2S is taken as an HCC chemotherapy curative effect prediction index, the potential of a new drug-resistant target is reversed, and related researchers are facilitated to improve the curative effect and improve the prognosis of patients by overcoming HCC chemotherapy drug resistance.
Owner:桂亮
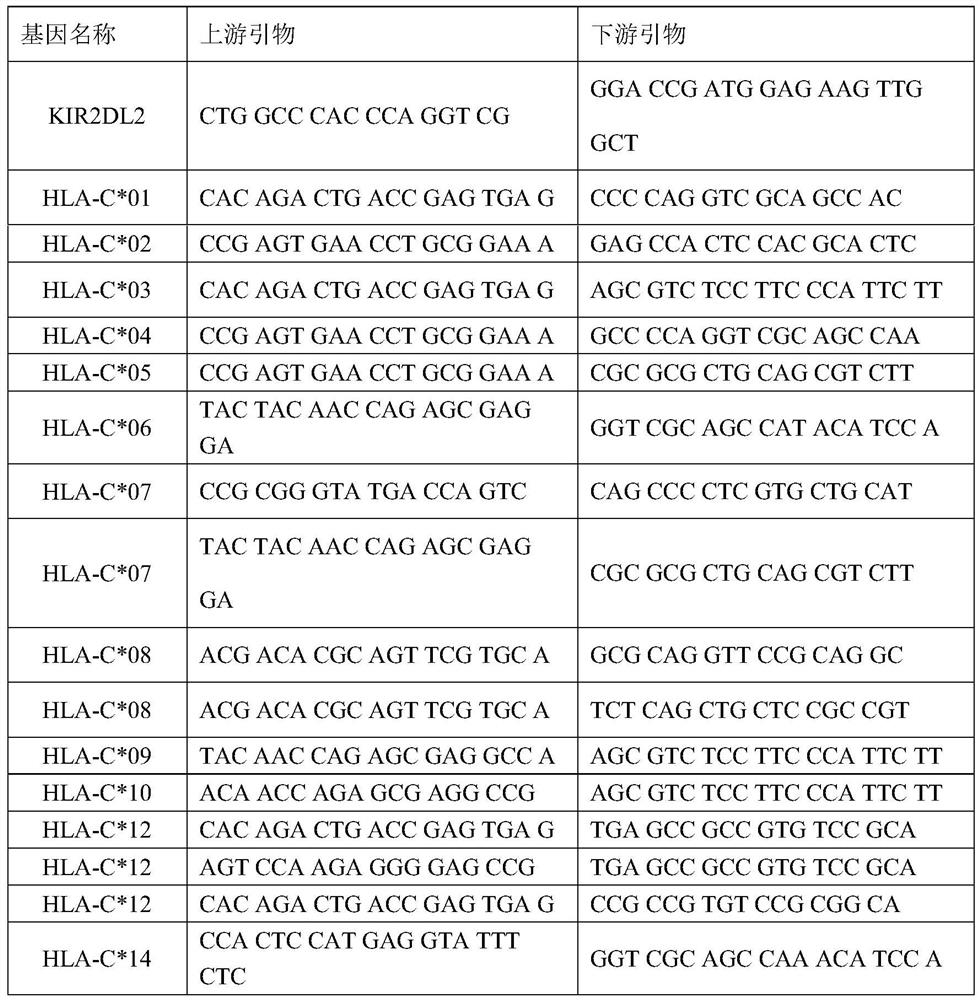
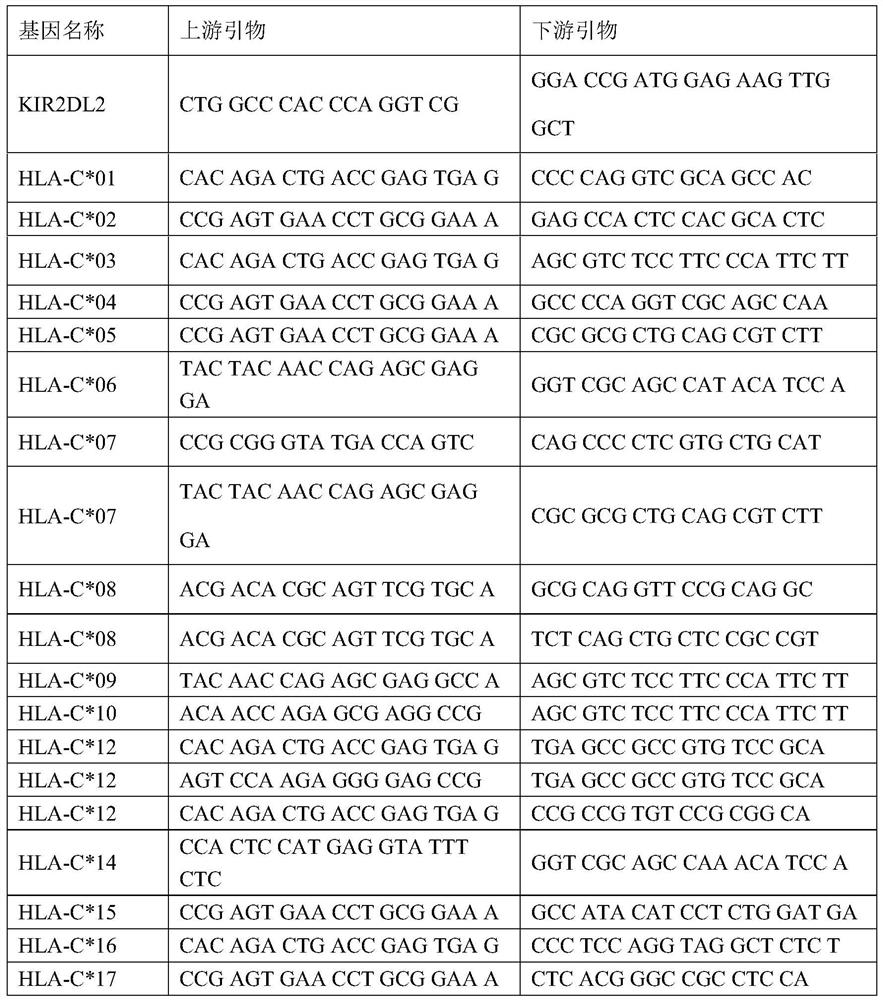
SARS-CoV-2 susceptible gene combination and application in preparation of COVID-19 susceptible population screening kit
The invention belongs to the field of medical detection, and particularly relates to an SARS-CoV-2 susceptible gene combination and application in preparation of a COVID-19 susceptible population screening kit. The human gene combination disclosed by the invention is KIR2DL2 and HLA-C, and specifically is HLA-C*01, HLA-C*02, HLA-C*03, HLA-C*04, HLA-C*05, HLA-C*06, HLA-C*07, HLA-C*08, HLA-C*09, HLA-C*10, HLA-C*12, HLA-C*14, HLA-C*15, HLA-C*16 and HLA-C*17. At present, early-stage prediction related researches on the occurrence of SARS-CoV-2 virus infection and susceptible populations do not exist in the world. The kit provided by the invention has great application value in disease susceptible population.
Owner:JIANGHAN UNIVERSITY
A quantitative method for carp spring viremia virus
ActiveCN109457016BSimple and fast operationLow costMicrobiological testing/measurementBiological testingAbsolute quantificationAssociated Study
The present invention amplifies the nucleoprotein gene N of SVCV through molecular biology techniques, and constructs recombinant plasmid pMD TM 18‑T‑SVCV‑N, using this plasmid as a template to establish an absolute quantitative SVCV copy number method based on the TaqMan probe method qPCR as a standard curve. At the same time, the protein concentration of SVCV was measured by Bradford method, and the correlation with the copy number of SVCV measured by the qPCR method was established. For the first time, a method for directly measuring protein concentration to determine the copy number of SVCV was established. This method can realize the simple and low-cost determination of the copy number of SVCV, and at the same time has important reference significance for the quantification of other viruses, and has important value in the related research of SVCV and other viruses.
Owner:SHENZHEN UNIV +1
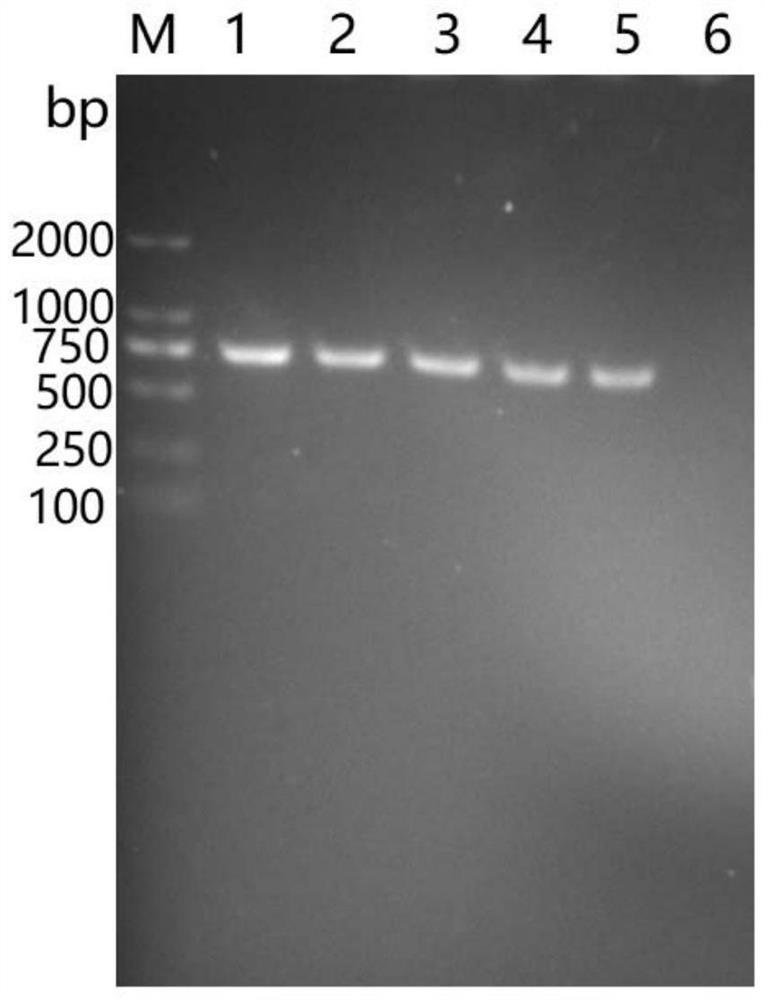
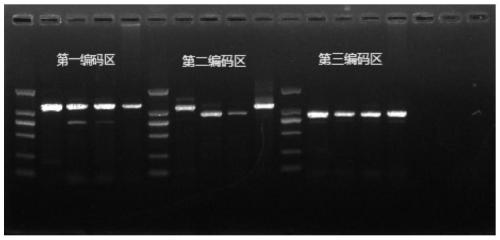
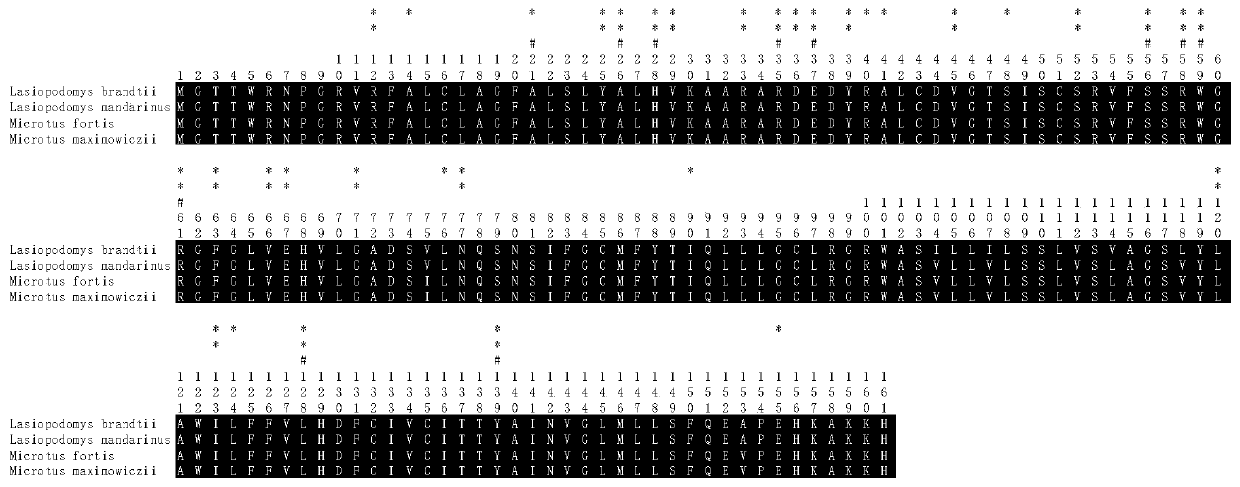
Primer Sets and Methods for Specific Amplification of the Coding Region of vkorc1 Genes of Voles and Microtus
The invention belongs to the field of biotechnology, particularly discloses a primer group and method for specific amplification of Cricetulus and Lasiopodomys Vkorc1 gene coding sequence region. Theprimer group includes 3 pairs of primers for respectively amplifying three exons of Vkorc1 genes, the primer sequence for amplifying the first exon is shown in SEQ ID NO.1-2, the primer sequence for amplifying the second exon is shown in SEQ ID NO.3-4, and the primer sequence for amplifying the third exon is shown in SEQ ID NO.5-6. The invention provides the amplification method based on the primer group through optimizing the amplification condition, the Cricetulus and Lasiopodomys Vkorc1 genes are amplified, and a good foundation is laid for previous related research on the Cricetulus and Lasiopodomys Vkorc1 genes and resistance to drugs.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
Preparation method and application of hawthorn EST-SSR (Expression Sequence Tag-Simple Sequence Repeats) marker primer
InactiveCN108330207APromote and improve the level of basic researchMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceBasic research
The invention discloses a preparation method of a hawthorn EST-SSR (Expression Sequence Tag-Simple Sequence Repeats) marker primer. The preparation method comprises the following steps: 1) screening and assembling segment sequences with the length greater than 5000bp from hawthorn transcriptome data; 2) carrying out SSR digging on each segment by utilizing SSR Hunter software; picking out EST sequences which contain rich dinucleotide, trinucleotide, tetranucleotide or pentanucleotide repeats and has the length greater than or equal to 10bp from a single sequence; 3) screening an EST-SSR primeron 150bp base sequences at upstream and downstream of SSR by utilizing primer 3 software, so as to obtain the hawthorn EST-SSR marker primer. The invention provides the preparation method and application of the hawthorn EST-SSR marker primer, reference is provided for related researches of hawthorns in the future and a basic research level of the hawthorns is promoted and improved.
Owner:SHENYANG AGRI UNIV
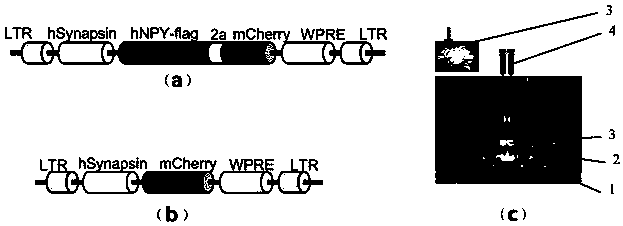
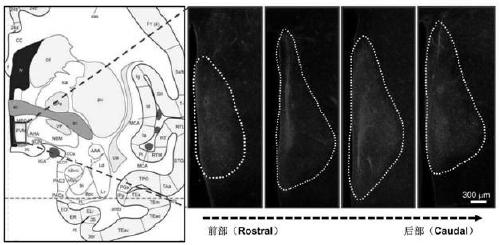
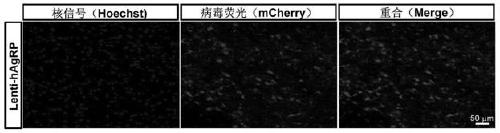
Construction method of gene overexpression chimeric animal model based on hAgRP and application of construction method
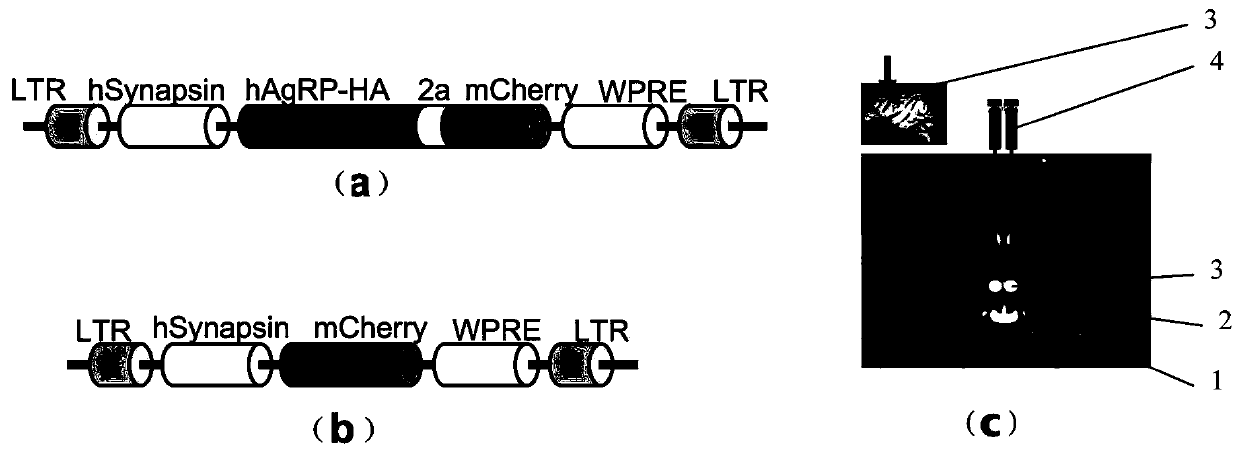
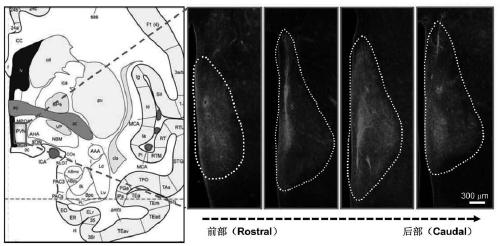
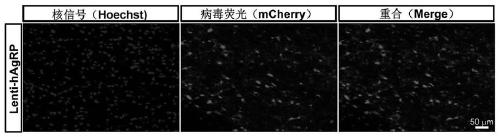
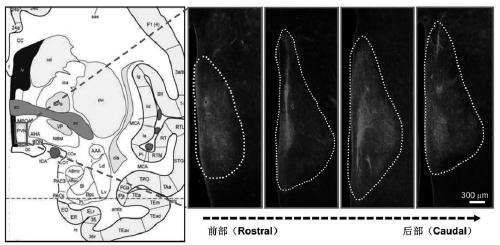
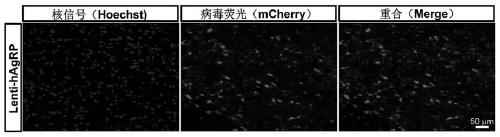
PendingCN111172193AShort build cycleImprove efficiencyDiagnostic recording/measuringSensorsDiseaseMacaque
The invention discloses a construction method of a gene overexpression chimeric animal model based on hAgRP and application of the construction method. The method comprises the following steps: takingconstructed Synapsin as a promoter to drive an hAgRP lentiviral expression vector; injecting the vector into the paraventricular nucleus of macaque; and transcribing hAgRP into paraventricular nucleus cell gene to realize overexpression of an hAgRP gene, and inducing phenotype generation to form the gene overexpression chimeric animal model based on the hAgRP. The method can be used for pathogenesis research of related diseases, screening, verification and testing of therapeutic drugs researched by clinical treatment means, pre-clinical related research and the like. The method has the characteristics of short animal model construction period, high efficiency, low cost, high phenotypic explicit rate, good synchronism, high repeatability, low difficulty, convenience in large-scale application and the like.
Owner:四川横竖生物科技股份有限公司
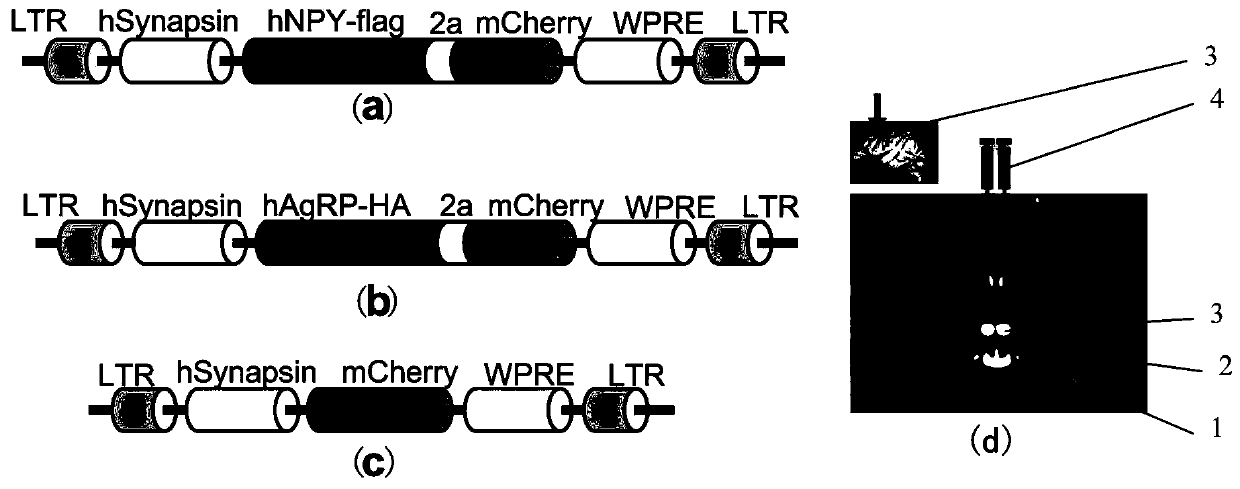
Construction method of gene overexpression chimeric animal model based on hNPY and hAgRP and application
PendingCN111172194AShort build cycleImprove efficiencyDiagnostic recording/measuringSensorsDiseasePromoter
The invention discloses a construction method of a gene overexpression chimeric animal model based on hNPY and hAgRP and application of the construction method. The method comprises the following steps: taking constructed Synapsin as a promoter to drive an hNPY and hAgRP lentiviral expression vector; injecting the vector into the paraventricular nucleus of macaque; and transcribing hNPY and hAgRPinto paraventricular nucleus cell gene to realize overexpression of hNPY and hAgRP genes, and inducing phenotype generation to form the gene overexpression chimeric animal model based on the hNPY andhAgRP. The method can be used for pathogenesis research of related diseases, screening, verification and testing of therapeutic drugs researched by clinical treatment means, pre-clinical related research and the like. The method has the characteristics of short animal model construction period, high efficiency, low cost, high phenotypic explicit rate, good synchronism, high repeatability, low difficulty, convenience in large-scale application and the like.
Owner:四川横竖生物科技股份有限公司
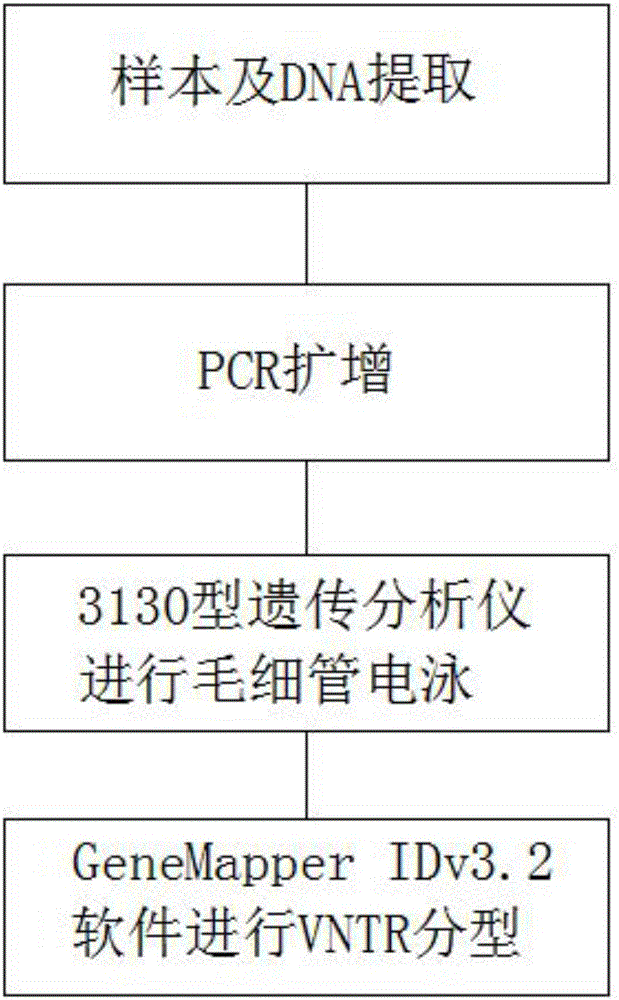
Automatic typing method of MAOAmu-VNTR fluorescent labels
InactiveCN106244711AImprove typing efficiencyEasy to handleMicrobiological testing/measurementCapillary electrophoresisElectrophoresis
The invention relates to the field of fluorescently-labeled primer typing, in particular to an automatic typing method of MAOAmu-VNTR fluorescent labels. The method comprises the steps of sample and DNA extraction, PCR amplification, capillary electrophoresis conducted through a 3130 type genetic analyzer and VNTR typing conduced through GeneMapper IDv3.2 software. The automatic typing method of the MAOAmu-VNTR fluorescent labels has the advantages that the typing efficiency of the built automatic typing method of the MAOAmu-VNTR polymorphic fluorescent labels is high, and by means of full-automatic capillary electrophoresis, MAOAmu-VNTR genotyping can be conducted more accurately; the advantages of being simple in sample treatment, high in degree of automation, short in detection time and the like are achieved, the result is accurate and reliable, operation is easy and convenient, and a good application prospect in related research for MAOAmu-VNTR polymorphism is achieved.
Owner:KUNMING MEDICAL UNIVERSITY
Construction method of gene overexpression chimeric animal model based on hNPY and application of construction method
PendingCN111172192AShort build cycleImprove efficiencyDiagnostic recording/measuringSensorsDiseaseMacaque
The invention discloses a construction method of a gene overexpression chimeric animal model based on hNPY and application of the construction method. The method comprises the following steps: takingconstructed Synapsin as a promoter to drive an hNPY lentiviral expression vector; injecting the vector into the paraventricular nucleus of macaque; and transcribing hNPY into paraventricular nucleus cell gene to realize overexpression of hNPY gene, and inducing phenotype generation to form the gene overexpression chimeric animal model based on the hNPY. The method can be used for pathogenesis research of related diseases, screening, verification and testing of therapeutic drugs researched by clinical treatment means, pre-clinical related research and the like. The method has the characteristics of short animal model construction period, high efficiency, low cost, high phenotypic explicit rate, good synchronism, high repeatability, low difficulty, convenience in large-scale application andthe like.
Owner:四川横竖生物科技股份有限公司
QRT-PCR reference genes of grapes and application of qRT-PCR reference genes
PendingCN112159864AMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyReference genes
The invention discloses qRT-PCR reference genes of grapes and an application of the qRT-PCR reference genes. The stability of ten candidate reference genes (Actin, 18s-rRNA, GAPDH, VvEF1-gamma, VvEF1-alpha, EF1-alpha, Ubiquitin, RRM1, PPR2 and MRE11) are analyzed in fruits, leaves and tendrils of seven different grape varieties and nine Jufeng fruit samples in different development stages throughthree pieces of analysis software including geNorm, NormFinder and BestKeeper according to early transcriptome data. Results show that the optimal reference gene number of the grape samples is two, the selection of the optimal reference genes of different samples is different, the optimal reference gene combination in the fruits comprises the VvEF1-gamma and the 18s-rRNA, the reference genes of the tendrils are the EF1-alpha and the Actin, and the reference genes in the fruit development stage are the EF1-alpha and the VvEF1-alpha. The qRT-PCR reference genes can provide important reference for related researchers to carry out qRT-PCR work in the grapes.
Owner:HENAN UNIV OF SCI & TECH
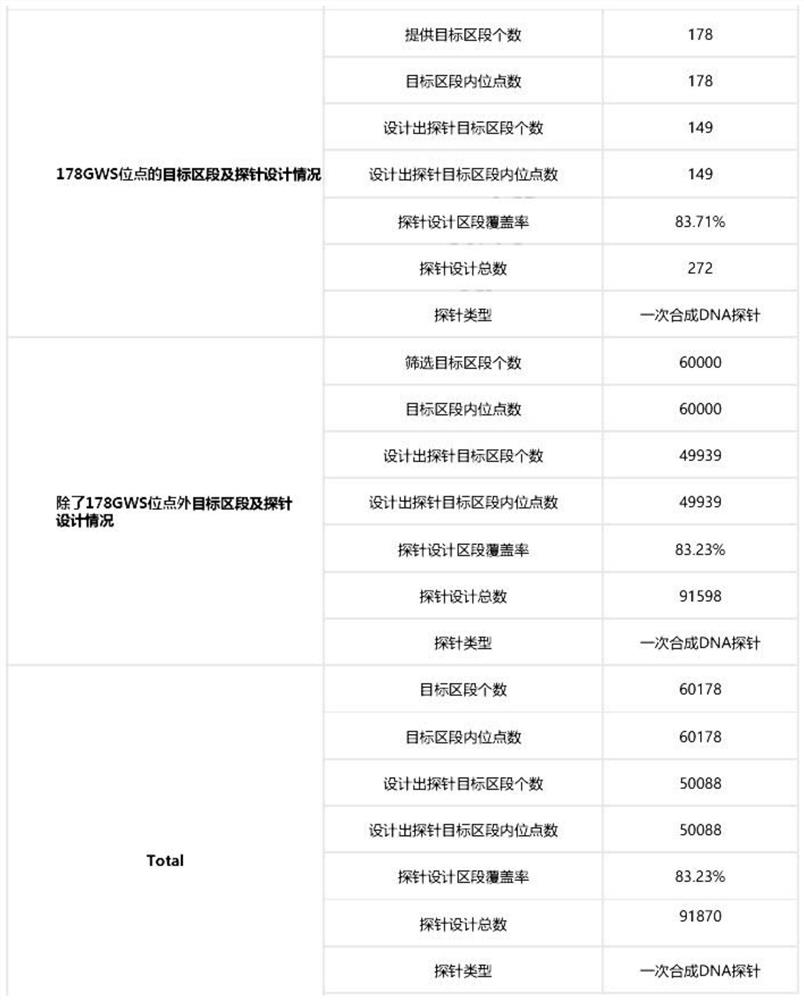
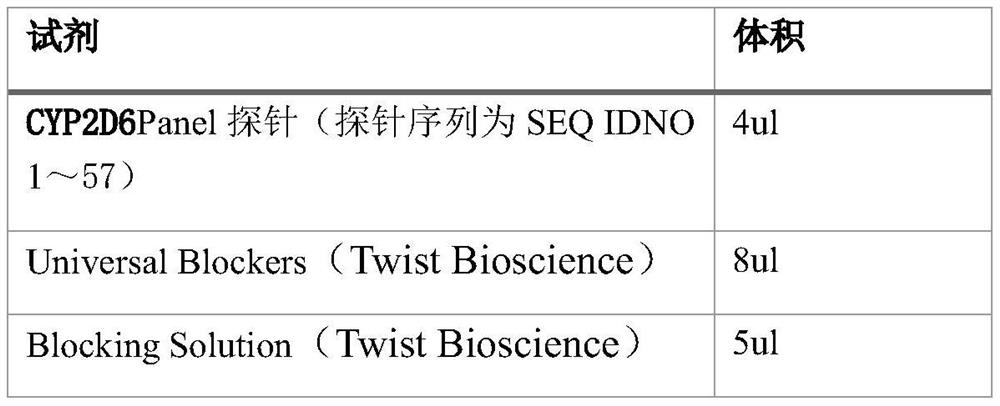
Group of probes and library building kit for detecting polymorphism of pharmacogenomics related gene CYP2D6 by using hybrid capture method
PendingCN114317533AShort timeSimple and fast operationMicrobiological testing/measurementLibrary creationGenomicsLarge sample
The invention provides a group of probes and a kit containing the group of probes. The group of probes and the kit containing the group of probes are used for manufacturing a library building kit for detecting polymorphism of a pharmacogenomics related gene CYP2D6 by using a hybrid capture method. According to the present invention, the CYP2D6 genotype and the drug metabolism capability can be rapidly obtained through the one-time detection, and the kit has advantages of simple operation, low time consumption, easy automatic detection, large sample size related research, and great significance on the discovery of the new genotype.
Owner:WUHAN ADICON CLINICAL LAB
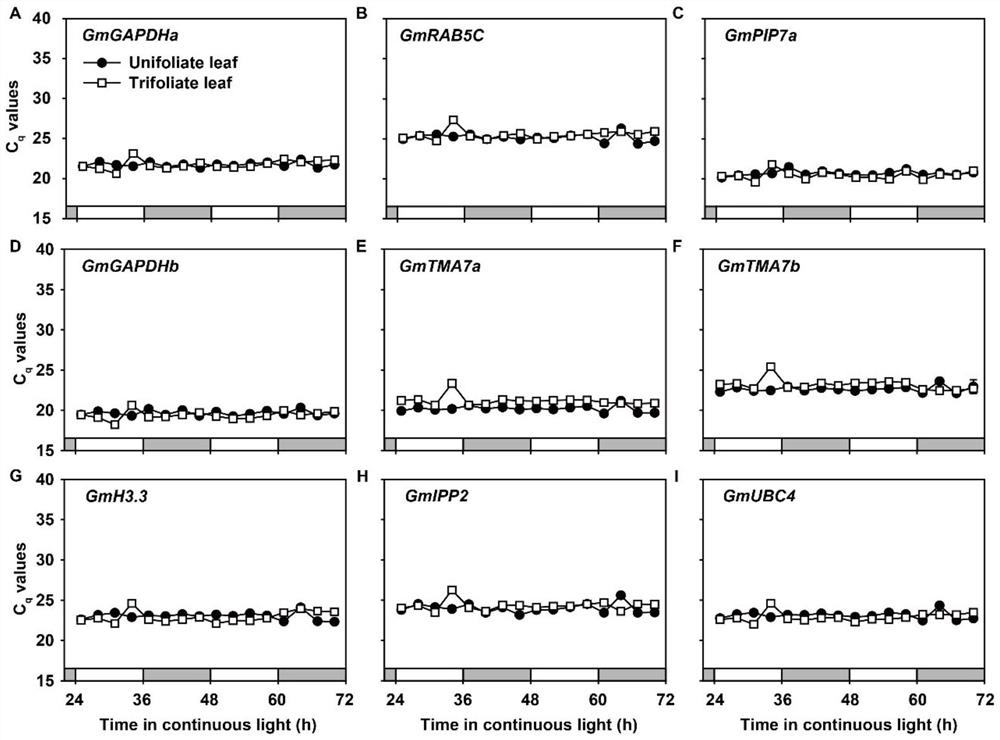
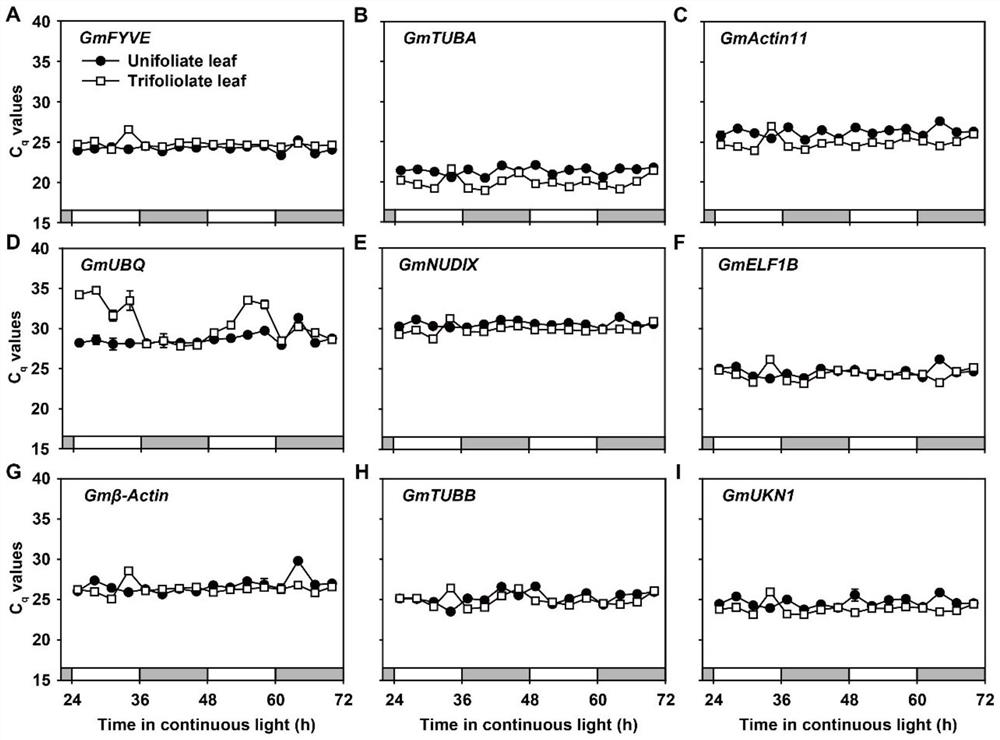
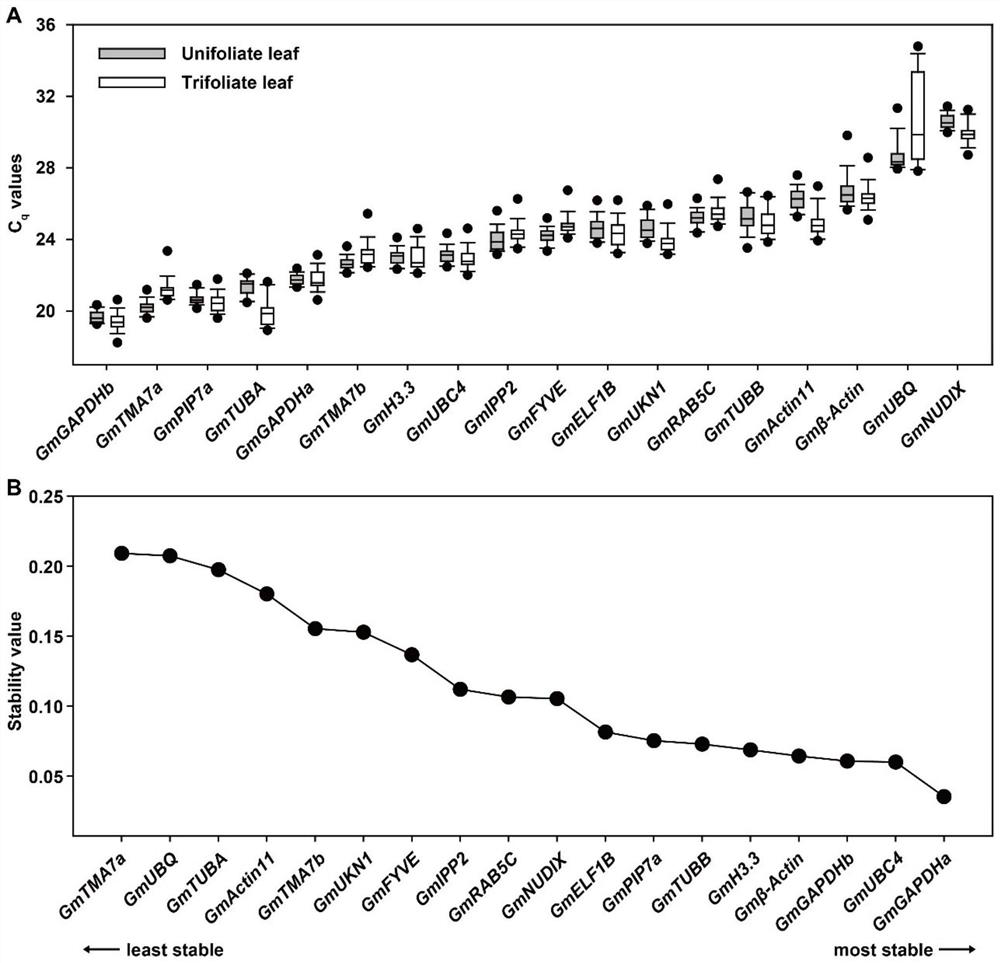
Soybean reference gene as well as detection primer and application thereof
The invention discloses a soybean reference gene as well as a detection primer and application thereof. The soybean reference gene is a group of reference genes used for analyzing gene recent rhythm expression, circadian rhythm expression and drought stress response in a single leaf and a compound leaf of soybeans, and comprises GmGAPDHa and GmRAB5C suitable for analyzing the three conditions in the single leaf and the compound leaf, and GmGAPDHb, GmH3.3 and GmPIP7a suitable for analyzing gene recent rhythm expression in the single leaf and the compound leaf. The reference gene provides favorable support for analyzing the stability and reliability of related research results of gene recent rhythm expression, circadian rhythm expression and drought stress response in soybean single leaf andcompound leaf, and particularly fills the blank of reference genes in soybean recent rhythm research.
Owner:HENAN UNIVERSITY
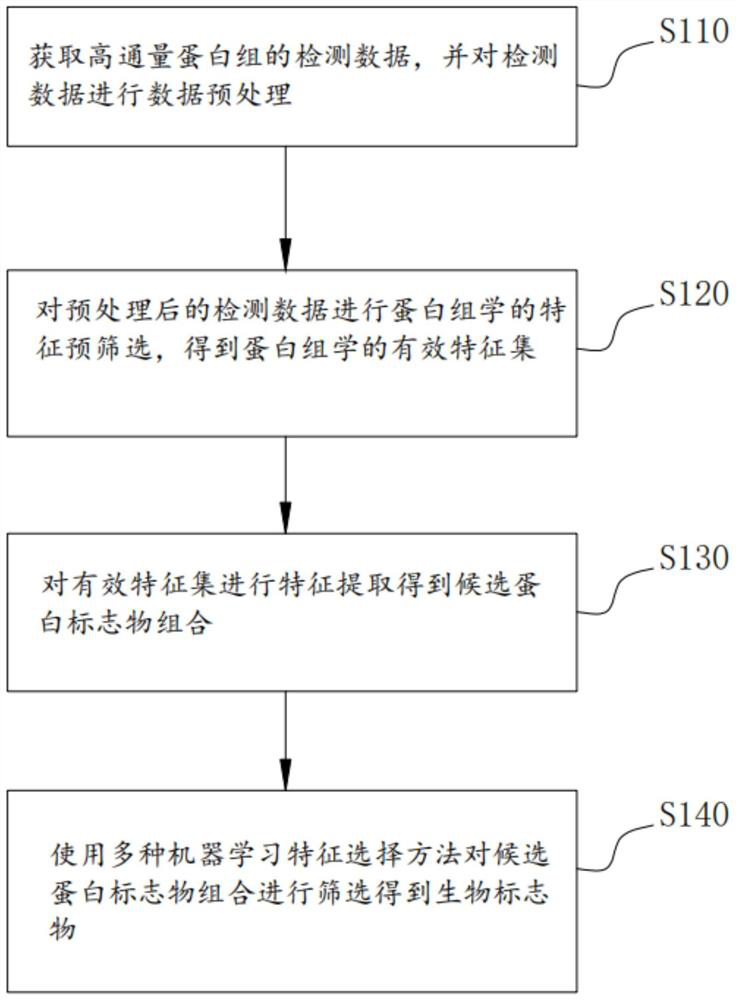
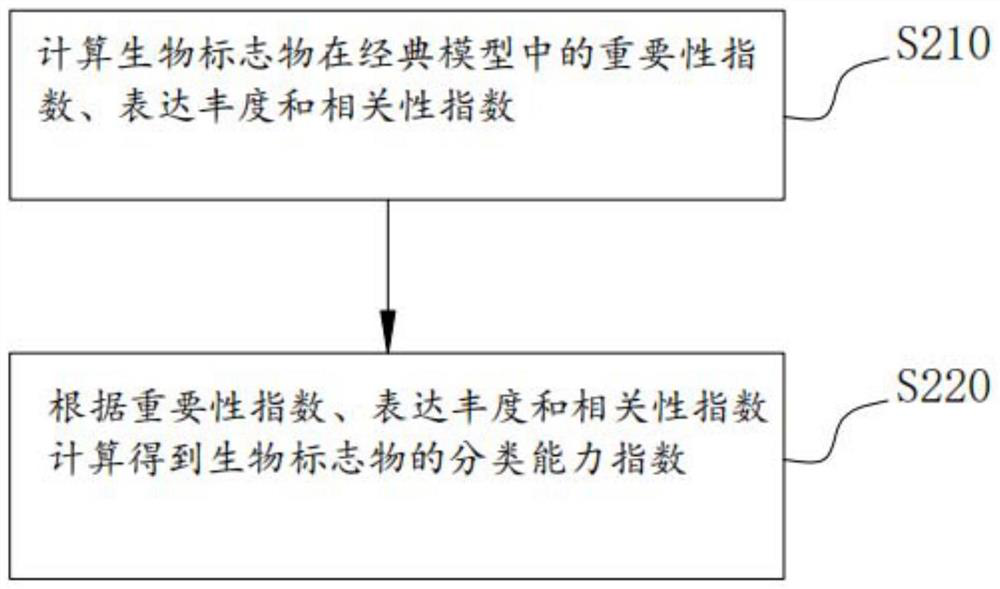
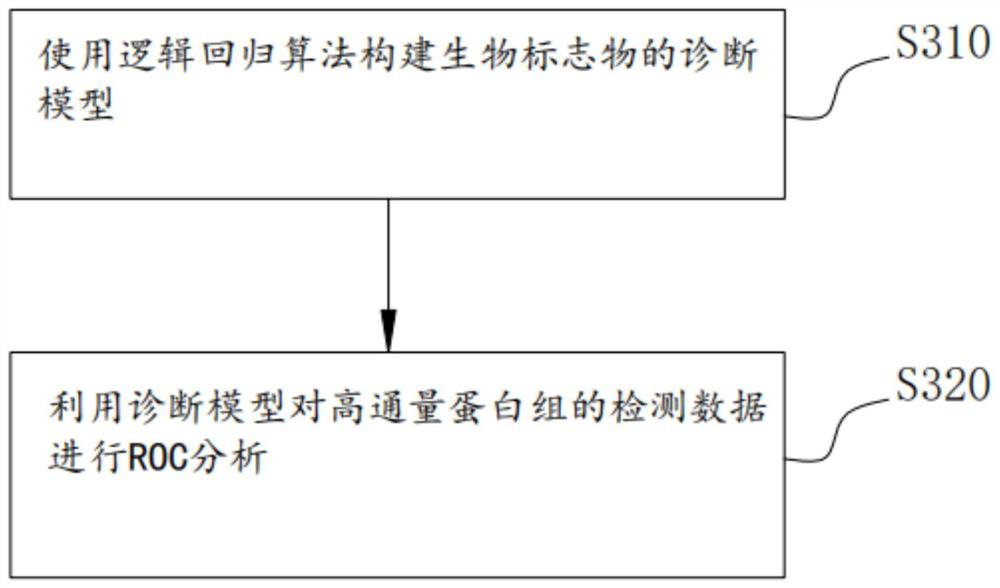
Method, system and medium for integrally screening proteome clinical biomarkers
PendingCN114550832AEasy to identifyEasy to classifyEnsemble learningBiostatisticsFeature setPotential biomarkers
The invention provides a proteome clinical biomarker overall screening method and system and a medium, and the method comprises the following steps: obtaining detection data of a high-throughput proteome, and carrying out data preprocessing on the detection data; carrying out proteomics feature pre-screening on the preprocessed detection data to obtain a proteomics effective feature set; performing feature extraction on the effective feature set to obtain a candidate protein marker combination; and screening the candidate protein marker combination by using a plurality of machine learning feature selection methods to obtain the biomarker. The method can be used for screening potential biomarkers with high throughput, high sensitivity, high accuracy and reasonable cost from mass data. The biomarker diagnosis with high verification rate and remarkable classification effect can be efficiently identified and identified, so that the time, energy and resources of related researchers can be greatly saved, and great convenience is provided for the related researchers.
Owner:WUHAN GENECREATE BIOLOGICAL ENG CO LTD
A method for screening disease-associated proteins based on complex networks
ActiveCN111640468BImprove accuracyReduce workloadBiostatisticsMachine learningDiseaseProtein Interaction Networks
The invention discloses a method for screening disease-related proteins based on a complex network. The method is as follows: 1) Obtain the seed gene related to the target disease; 3) extracting the feature data of the protein in the protein interaction network; 4) using the feature data of the protein as training data, and using a machine learning algorithm to train a PU classifier; 5) predicting the PU classifier according to the PU classifier Proteins in the protein interaction network associated with the target disease. The method of the invention can quickly and efficiently identify proteins related to diseases, and is helpful for biomedical experts to carry out experimental verification or relevant researchers to carry out work.
Owner:天士力国际基因网络药物创新中心有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com