Li SSR marker primer pair developed based on transcriptome sequence and its application
A technology of transcriptome sequence and labeling primers, which is applied in recombinant DNA technology, microbial determination/inspection, biochemical equipment and methods, etc., to achieve the effect of increasing original data and improving the efficiency of development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
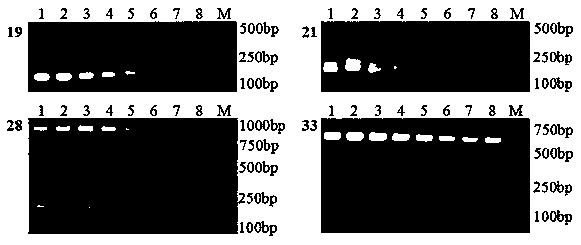
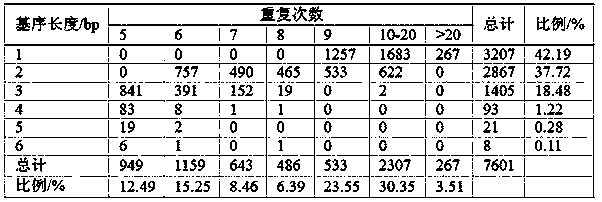
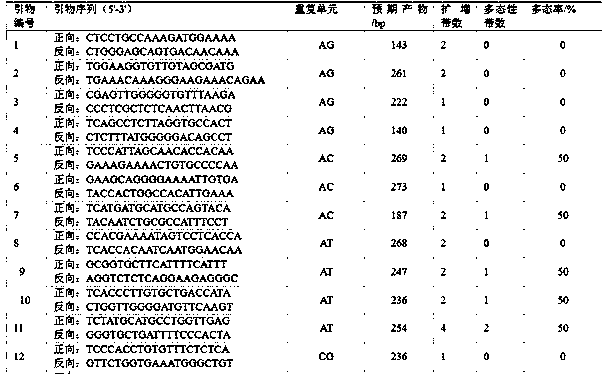
[0058] The plum SSR primer pair developed based on transcriptome sequencing is obtained through plum transcriptome sequencing, SSR site identification, SSR primer design, DNA extraction, SSR primer screening, and genetic diversity analysis.
[0059] In the present invention, the specific method of the plum SSR primer pair developed based on the transcriptome sequence is:
[0060] 1) Acquisition of transcriptome data
[0061] EZNA Plant RNA Kit (Omega Bio-tek) was used for the extraction of total RNA from hibiscus plum fruit. The RNA concentration was determined by Qubit 2.0 Fluorometer (Invitrogen, Life Technologies, CA, USA), and the quality of RNA was evaluated by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The RNA from the fruit samples of the same stage of three different trees was mixed in equal amounts and used for sequencing library construction.
[0062] The EZNA Plant RNA Kit (Omega Bio-tek) was used to extract the total RNA from the hibiscus plum ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com