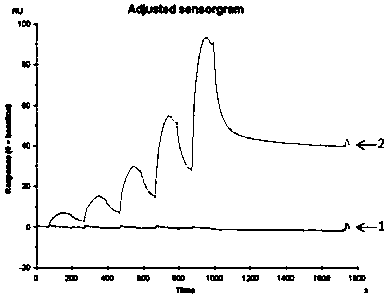
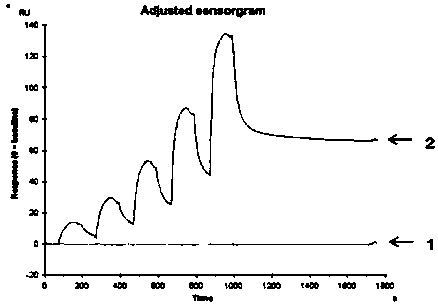
A single domain antibody that recognizes hla-a2/rmfpnapyl
A technology for HLA-A2 and single-domain antibodies, which is applied in the field of single-domain antibodies that recognize HLA-A2/RMFPNAPYL, and can solve the problems of limited literature
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Screening the single domain antibody of HLA-A2 / RMFPNAPYL complex
[0052] 1.1 Single domain antibody phage library preparation
[0053] 1.1.1 Preparation of helper phage (BM13)
[0054] The M13KE phage replicon was double-digested with AlwnI and AfeI, and the artificially synthesized gene fragment was also double-digested with AlwnI and AfeI, and then ligated together with T4 ligase. After ligation, TG1 was transfected to obtain helper phage BM13. Thus, in the protoreplicon
[0055] The tctggtggtggttctggtggcggctctgagggtggtggctctgagggtggcggttctgagggtggcggctctgagggaggcggttccggtggtggctct sequence was replaced by a synthetic gene sequence, that is, a trypsin cleavage sequence was added in the phage GIII coding region. Once used as a helper phage, the trypsin digestion step was added to reduce the number of phages that did not contain the fusion target gene protein.
[0056] The synthetic gene sequence is as follows:
[0057] CCA GCC GGC CTT TCT GAG GGG TCG ACT...
Embodiment 2
[0102] Example 2, Expression of Single Domain Antibody
[0103] Prokaryotic single domain antibody expression
[0104] The four single domain antibody genes were cloned into pET22b with Nco I and Not I restriction endonucleases to express monovalent antibodies. The constructed vector was transformed into E.coli / DE3, single clones were picked the next day, and cultured with shaking at 220 rpm at 37°C until the OD600 was about 0.5. After adding IPTG (working concentration: 1 mM), the expression was induced at 220 rpm at 18°C for 20 hours. Detection of protein expression. The sonication supernatant was purified with ProteinA and run SDS-PAG, see Figure 4 . Among them, the Marker strips are 14, 25, 30, 40, 50, 70, 100, 120, 160KD from small to large. Line1 is M5-H1, line2 is M5-G3, and line3 is M5-F4. Single domain antibodies are about 14KD in size.
[0105] Fusion FC expression in pET22b In order to form a bivalent antibody, the single domain antibody gene was connected ...
Embodiment 3
[0112] Example 3. Specific recognition of HLA-A2 / RMFPNAPYL complex by single domain antibody
[0113] Specific recognition of antigen complexes by single domain antibodies
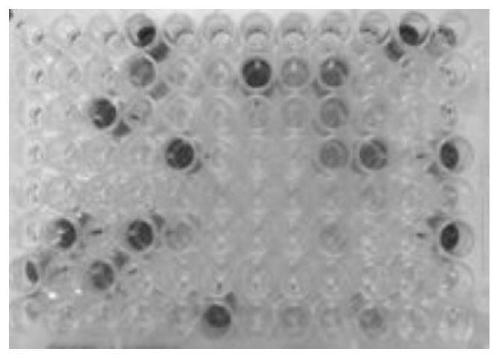
[0114] For the single-domain antibody expressed on pET22b, the cells were collected, resuspended evenly in PBS, and then sonicated. Ultrasonic crushing conditions: 600W, ultrasonic for 2 seconds, interval of 6 seconds, 10 minutes in total, 16°C. After ultrasonication, centrifuge at 12000rpm at 4°C for 10 minutes, take the supernatant for ELISA identification of the specificity of different antigens (Protein A-HRP is the secondary antibody) and read the plate. For data analysis, see Figure 9 . Wherein, the ordinate is the light absorption value at 650nm, and the abscissas 1, 2, 3, 4 are the four antigens HLA-A2 / ITDQVPFSV, HLA-A2 / NLVPMVATV, HLA-A2 / RMFPNAPYL, HLA-A2 / SLLMWITQC. The three samples are the ultrasonic supernatants of M5-H1, M5-G3, and M5-F4 expressed separately in pET22b.
[0115] The results...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com