A multi-trait and multi-interval localization method based on genotype error at marker loci
A technology for marking sites and genes, applied in the field of molecular biology, can solve problems such as information errors of marker genes, and achieve the effect of cost saving and accurate positioning
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0032] The present invention provides an embodiment of a multi-trait multi-interval positioning method based on marker locus genotypes containing errors, in order to enable those skilled in the art to better understand the technical solutions in the embodiments of the present invention, and make the present invention The above objects, features and advantages can be more obvious and understandable, and the technical solution in the present invention will be further described in detail below in conjunction with the accompanying drawings:
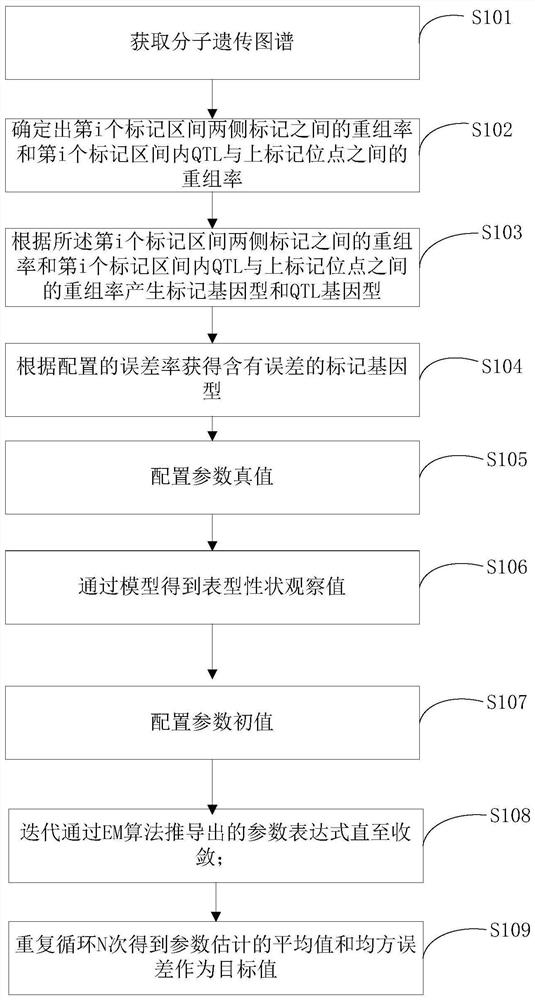
[0033] The present invention firstly provides a multi-trait multi-interval positioning method based on marker locus genotype containing errors, such as figure 1 shown, including:
[0034] S101, step 1, obtaining a molecular genetic map;
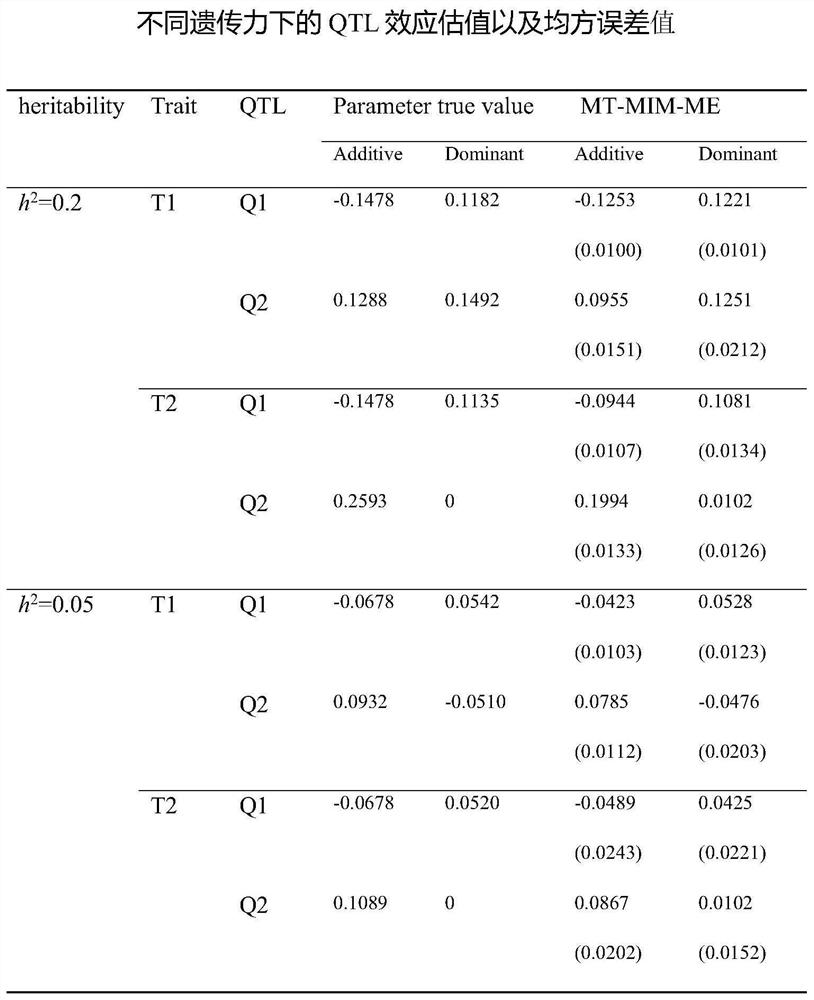
[0035] The data in this example comes from the work of Wittenburg et al. 2003, and this data set contains 305 F2 offspring. For the phenotype data, gallstone weight and high-density lipoprotein (High-densi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com