Application of miR-577 for preparing nephrosis diagnosis marker
A technology of mir-577 and diagnostic reagents, which is applied in the determination/testing of microorganisms, drug combinations, biochemical equipment and methods, etc., and can solve the problems of low early diagnosis rate, lost operation opportunities, hidden onset of renal clear cell carcinoma, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1 TCGA data analysis
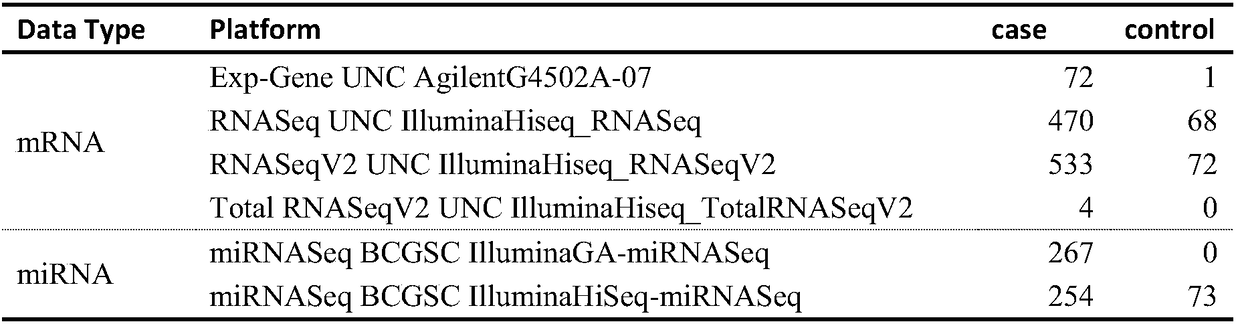
[0047] The mRNA and miRNA data generated by the TCGA database were used. These data were obtained from Primary solid Tumor in the renal clear cell carcinoma case group, and Solid Tissue Normal or Control Analyte* in the control group. The search results are detailed in Table 1 below.
[0048] Table 1
[0049]
[0050] mRNA data analysis results: TCGA database IlluminaHiseq_RNASeqV2 platform has a total of 20531 genes. In this study, 7950 genes that were not easily detected were deleted (only genes without 0 elements in the data were kept). 12581 genes were used for screening of differentially expressed genes. The indicators for screening significantly different mRNAs are: p_value1, |value1-value2|>100, 2361 differentially expressed mRNAs were obtained, 1350 were up-regulated, and 1011 were down-regulated.
[0051] miRNA data analysis results: TCGA database IlluminaHiSeq-miRNASeq platform has a total of 1046 miRNAs. In this stud...
Embodiment 2
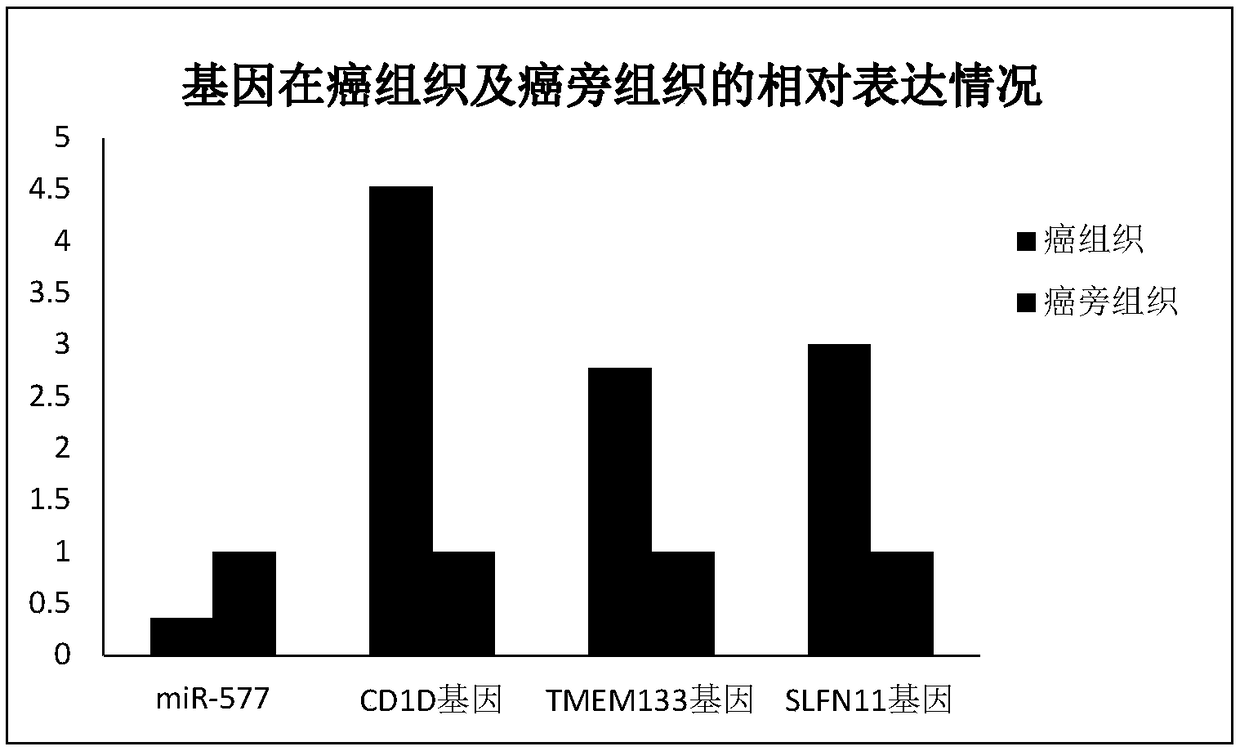
[0054] Example 2 Detection of expression of miR-577 and its target genes in clear cell renal cell carcinoma and adjacent tissues by real-time quantitative PCR
[0055] 2.1 Sample acquisition:
[0056] The cancer tissues and paracancerous tissues of 34 patients with clear cell renal cell carcinoma were selected, and all the above specimens were obtained with the consent of the organizational ethics committee.
[0057] 2.2 Extraction of total RNA from samples:
[0058] 1) Take 80 mg tissue block, add 800 μl Lysis / Binding buffer, and use a homogenizer to homogenize the tissue block. During the homogenization process, the samples should be placed on ice to keep the cold state.
[0059] 2) Add 1 / 10 volume of Homogenate Additive to the above homogenized tissue samples, and place on ice for 10 minutes.
[0060] 3) Add an equal volume of water-saturated phenol to the Lysis / Binding buffer, shake for 45 seconds, and centrifuge at 10,000×g for 5 minutes at room temperature.
[0061] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com