Substances for regulating chromatin histone methylation level of rDNA gene and application thereof
A technology of chromatin and trimethylation, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
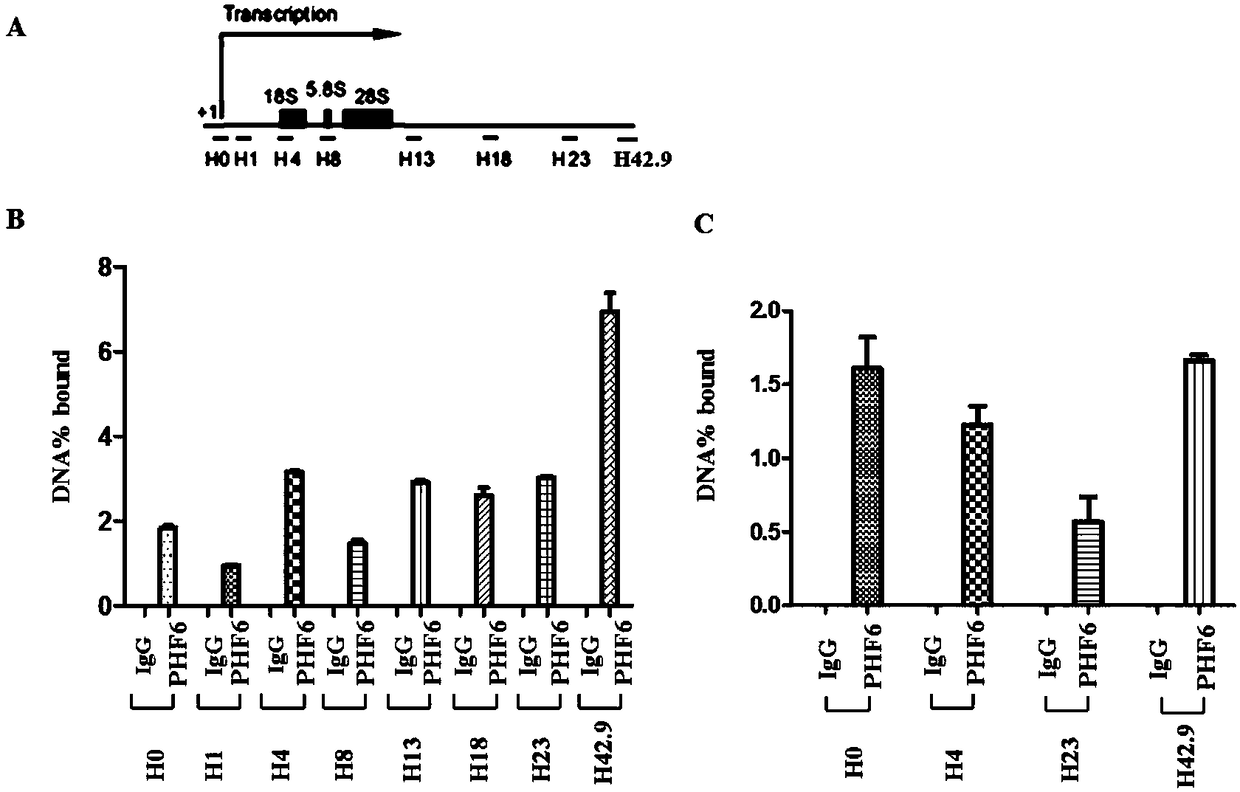
[0032] Example 1. PHF6 binds to the pre-rRNA transcription region of rDNA
[0033] 1. Design detection primers according to the sequence of the pre-rRNA transcription region on rDNA, a total of 8 pairs of primers, namely H0, H1, H4, H8, H13, H18, H23, H42.9, spread throughout the pre-rRNA transcription region ( figure 1 A).
[0034] H0 primer sequences are SEQ ID No.4 and SEQ ID No.5
[0035] F: GGTATATCTTTCGCTCCGAG SEQ ID No.4
[0036] R:GACGACAGGTCGCCAGAGGA SEQ ID No.5
[0037] H1 primer sequences are SEQ ID No.6 and SEQ ID No7 respectively
[0038] F: GGCGGTTTGAGTGAGACGAGA SEQ ID No.6
[0039] R: ACGTGCGCTCACCGAGAGCAG SEQ ID No7
[0040] H4 primer sequences are SEQ ID No.8 and SEQ ID No.9 respectively
[0041] F: CGACGACCCATTCGAACGTCT SEQ ID No.8
[0042] R:CTCTCCGGAATCGAACCCTGA SEQ ID No.9
[0043] H8 primer sequences are SEQ ID No.10 and SEQ ID No.11 respectively
[0044] F:AGTCGGGTTGCTTGGGAATGC SEQ ID No.10
[0045] R:CCCTTACGGTACTTGTTGACT SEQ ID No.11
[0046...
Embodiment 2
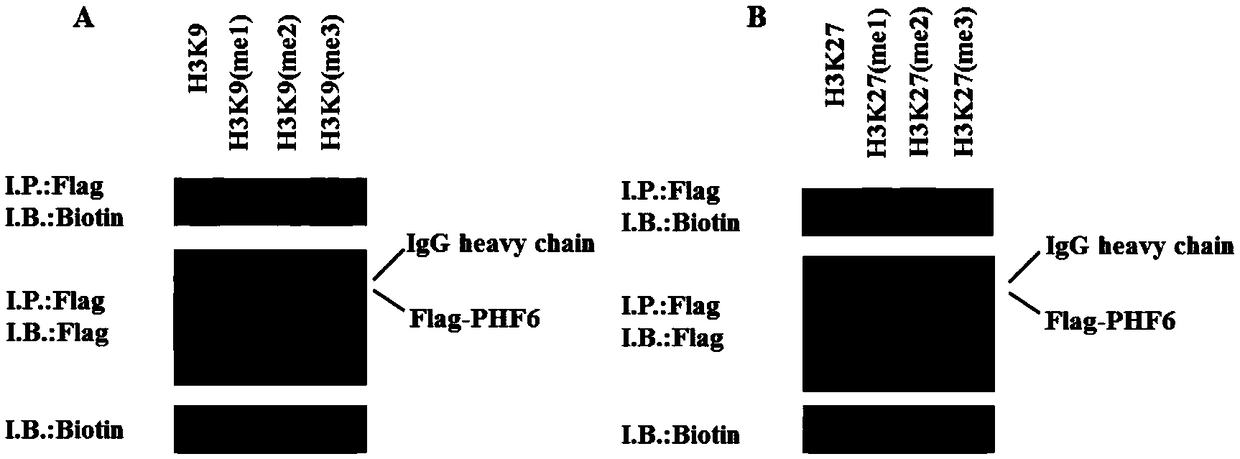
[0062] Example 2, PHF6 can bind to histone H3 methylation mimic peptide
[0063]Detection was performed by co-immunoprecipitation and immunoblotting. Artificially synthesized some histone H3 methylation modification site mimic peptides, including H3K9, H3K9me1, H3K9me2, H3K9me3, H3K27, H3K27me1, H3K27me2 and H3K27me3, these mimic peptides have biotin (Biotin) tags, can use antibiotics Primary antibody for detection. Transfer plasmid Flag-PHF6 into HEK 293 cells to express Flag-PHF6 protein, extract cell lysate after 48h, add anti-Flag primary antibody and synthetic H3K9, H3K9me1, H3K9me2, H3K9me3, H3K27, H3K27me1, H3K27me2 and H3K27me3 mimic peptides After co-incubating overnight, the Flag-antibody conjugates were precipitated with proteinA / G beads, and then analyzed by Western blotting. The experimental results show that Flag-PHF6 binds to H3K9me1 / H3K9me2 / H3K9me3, among which the binding to H3K9me1 and H3K9me2 is weak, and the binding of PHF6 to H3K9me3 is more ( figure 2...
Embodiment 3
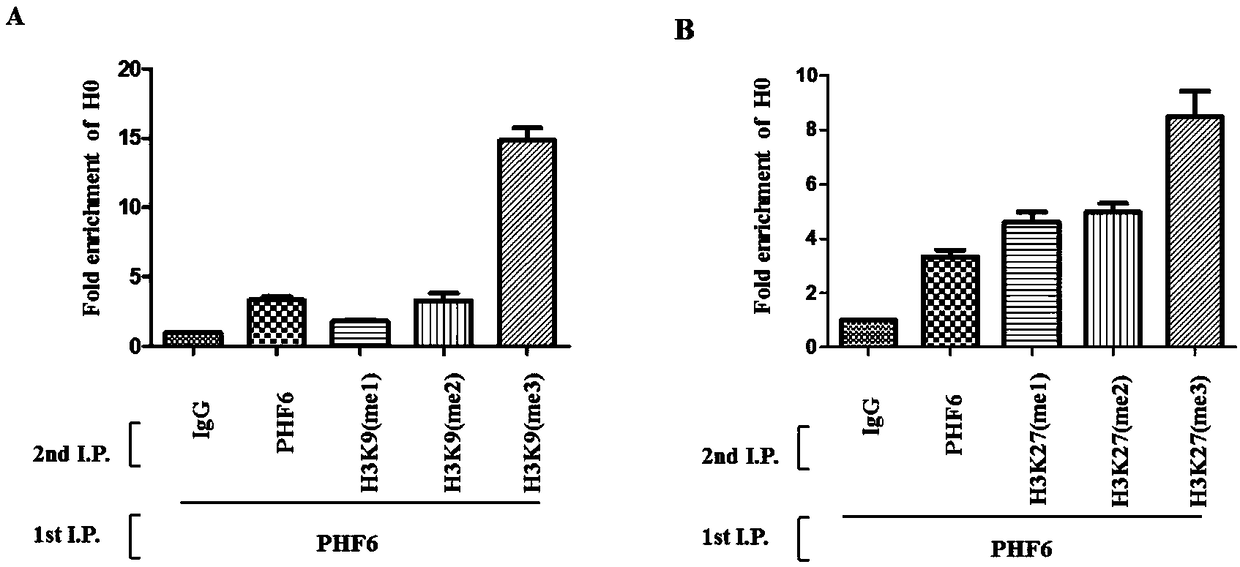
[0064] Example 3, PHF6 can bind to the H3K9 (me1 / 2 / 3) site and H3K27 (me1 / 2 / 3) site in the rDNA transcription initiation region
[0065] HeLa cells were selected and the experiment was carried out using the Re-ChIP method, which required two chromatin immunoprecipitation steps. First, collect the cell lysate after fixation with formaldehyde, collect the supernatant after sonication and centrifugation, incubate the supernatant with the primary antibody against PHF6, and add magnetic beads to obtain the small DNA fragment bound by the primary antibody against PHF6; Add 10mM dithiothreitol (DL-Dithiothreitol, DTT) to the magnetic beads bound to the small fragments of DNA bound by the primary antibody against PHF6 for elution operation, divide the eluate into the required aliquots, add cell lysis After diluting to an appropriate volume, the corresponding negative control IgG and anti-PHF6 / H3K9me1 / H3K9me2 / H3K9me3 / H3K27me1 / H3K27me2 / H3K27me3 primary antibodies were added to carry out...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com