A Dunaliella salina chloroplast homologous recombination empty vector and its application
A Dunaliella salina, homologous recombination technology, applied in the field of genetic engineering, can solve the problems of restricting basic research and application development, lack of high-efficiency and stable endogenous high-efficiency regulatory sequences at chloroplast insertion sites, and achieve high-efficiency endogenous regulatory sequences Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Cloning of Dunaliella salina chloroplast homologous recombination fragment
[0034] Design and synthesize the following two pairs of primers:
[0035] P1: 5'-TTACCAGGGTTTGACATGTCTAGAA-3'
[0036] P2: 5'-TGGGCTATAGAAGATTTGAAC-3'
[0037] P3: 5'-GGGAATGTAGCTCAGTTGGTAGAGC-3'
[0038] P4: 5'-TTCAGCTGTTTCGTTTTTAGAAAACT-3'
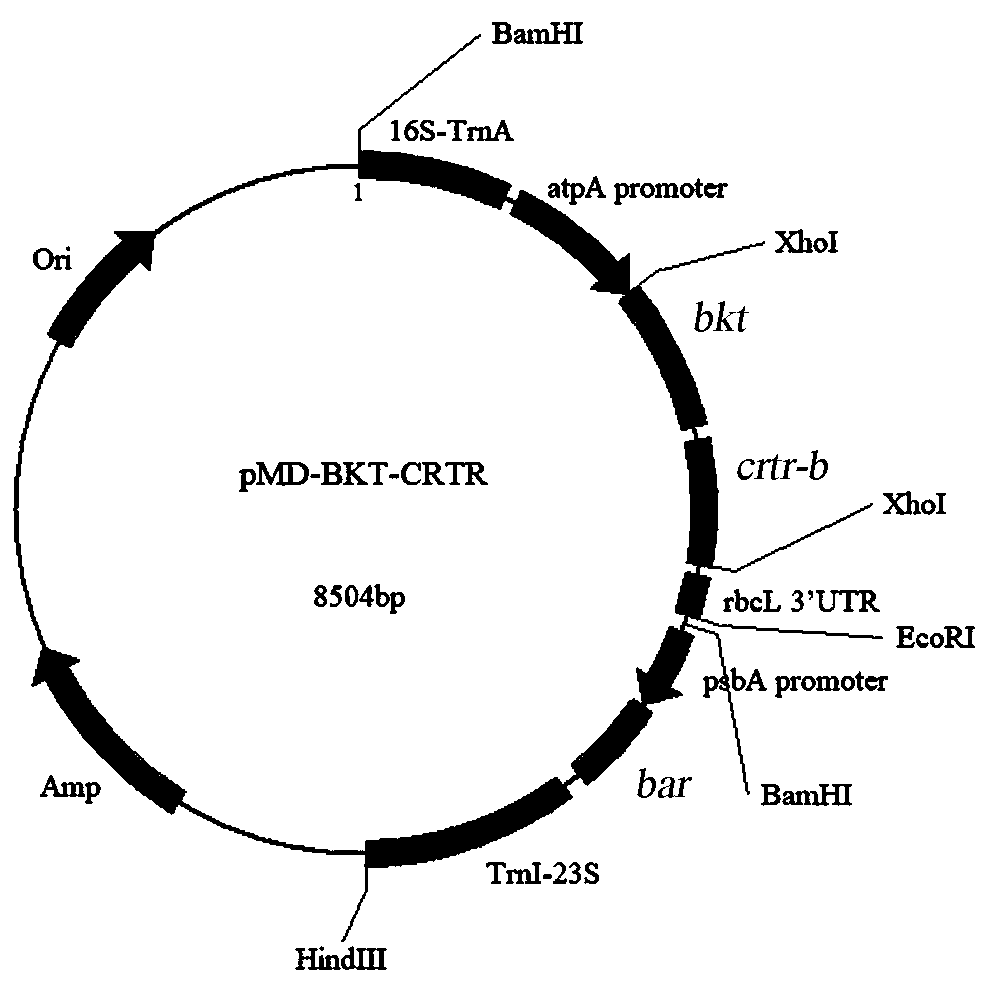
[0039] Wherein the amplified product of primers P1 and P2 is SEQ ID NO: 1, i.e. fragment 16S-TrnA; the amplified product of primers P3 and P4 is SEQ ID NO: 2, i.e. fragment TrnI-23S (see figure 1 ).
[0040] Using the total genome DNA of Dunaliella salina as a template, carry out PCR amplification with primers P1 and P2, the reaction program is: 94°C 10min pre-denaturation; 94°C 1min, 60°C 90sec, 72°C 90sec, a total of 30 cycles; 72°C 5min extend. The PCR amplification product is about 901bp, which is the fragment 16S-trnI. After the fragment is subjected to agarose gel electrophoresis, the PCR product purified by gel recovery (Tiangen Co...
Embodiment 2
[0042] Example 2 Amplification and cloning of two chloroplast promoter fragments of Dunaliella salina
[0043] Design and synthesize the following two pairs of primers:
[0044] P5: 5'-ATCCGCGTAGAGTAATAGG-3'
[0045] P6: 5'-GAGCACCATTTTTACTTCTGGTGTA-3'
[0046] P7: 5'-GGATCCGCCGATCCGTGGTTTAGAGTT-3'
[0047] P8: 5'-ACGTGCCCAAAGGCTAGTATTT-3'
[0048] Wherein the amplified product of primers P5 and P6 is SEQ ID NO: 3, that is, fragment 5'atpA, which is a promoter with chloroplast activation function derived from Dunaliella salina chloroplast; the amplified product of primers P7 and P8 is SEQ ID NO : 4, namely the fragment 5' psbA, is derived from the promoter with chloroplast activation function of Dunaliella salina chloroplast (see figure 2 ).
[0049] Using the total genome DNA of Dunaliella salina as a template, PCR amplification was carried out with primers P5 and P6. The reaction program was: 94°C 10min pre-denaturation; 94°C 1min, 60°C 90sec, 72°C 90sec, a total of 30 c...
Embodiment 3
[0051] Example 3 Construction of Dunaliella salina chloroplast homologous recombination empty vector
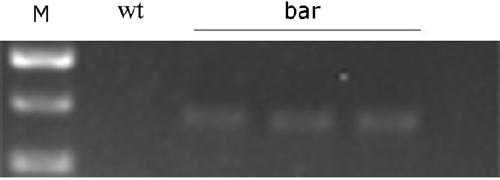
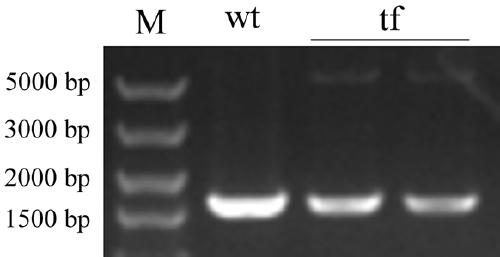
[0052] Based on the above-mentioned cloning vectors pMD16I, pMD23A, pMDatpA, and pMDpsbA, a Dunaliella salina chloroplast homologous recombination vector was constructed by means of homologous recombination.
[0053] Design and synthesize the following six pairs of primers:
[0054] P9: tagcctttgggcacgtATGAGCCCAGAACGACGCC
[0055] P10: ctgagctacattcccTCATCAAATCTCGGTGACGGG
[0056] P15: tgctcctcgagCTGCTTGTGAAGTTTGGAAAGAAA
[0057] P16: tgctcctcgagCTGCTTGTGAAGTTTGGAAAGAAA
[0058] P17: catgattacgaattcggatccTTACCAGGGTTTGACATGTCTAGAA
[0059] P18: gcggatTGGGCTATAGAAGATTTGAAC
[0060] P19: gatgaGGGAATGTAGCTCAGTTGGTAGAGC
[0061] P20: acgacggccagtgccaagcttTTCAGCTGTTTCGTTTTTAGAAAACT
[0062] P21: tatagcccaATCCGCGTAGAGTAATAGG
[0063] P22: aagcagctcgagGAGCACCATTTTTACTTCTGGTGTA
[0064] P23: tatgaccatgattacgaattcGGATCCGCCGATCCGTGGTTTAGAGTT
[0065] P24: tcatACGTGCCCAAAGGCTAG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com