Cellobiose epimerase mutant and application thereof in galactopoiesis fructose
A technology of epimerase and cellobiose, applied in the field of genetic engineering, can solve problems such as doubtful safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Recombinant plasmid pBSMuL3-ce (-)build process
[0035] The cellobiose epimerase (NCBI: WP_012547288.1) gene derived from thermophilic anaerobic bacteria was synthesized, and the gene fragment was connected to the pET-24a vector to obtain the plasmid pET-24a-ce. Using the plasmid pET-24a-ce as a template, the forward and reverse primers were designed as F1 / R1 (sequence information shown in SEQ ID NO.3 and SEQ ID NO.4, respectively) to amplify the CE gene fragment. The target fragment ce obtained by PCR was ligated with the pMD-19T Simple vector and transformed into E.coli JM109, and the single-clonal transformant was selected and cultured in LB liquid medium for 8-10 hours, the plasmid was extracted, and the double-enzyme digestion was verified to perform gene sequencing. The correctly sequenced plasmid was designated as pMD-19T-ce. Plasmid pMD-19T-ce was double digested with restriction endonucleases HindⅢ and BamHI, and the ce fragment was obtained by ge...
Embodiment 2
[0036] Embodiment 2: Recombinant plasmid pBSMuL3-ce (-) transformation
[0037] Plasmid pBSMuL3-ce (-) Transformed into Bacillus subtilis CCTCC NO:M2016536 prepared in advance to obtain genetically engineered bacteria Bacillus subtilis CCTCC NO:M 2016536 / pBSMuL3-ce (-) , spread on LB plates containing kanamycin (100 μg / mL) resistance, and incubate at 37°C for 10-12h. Pick a single colony into 10 mL of liquid LB medium containing kanamycin (100 μg / mL) resistance, culture at 37 ° C for 8 h, save the glycerol tube, and store it in a -80 ° C refrigerator. After the verification is correct, shake flask fermentation is carried out to produce enzyme.
Embodiment 3
[0038] Embodiment 3: Shake flask fermentation produces enzyme
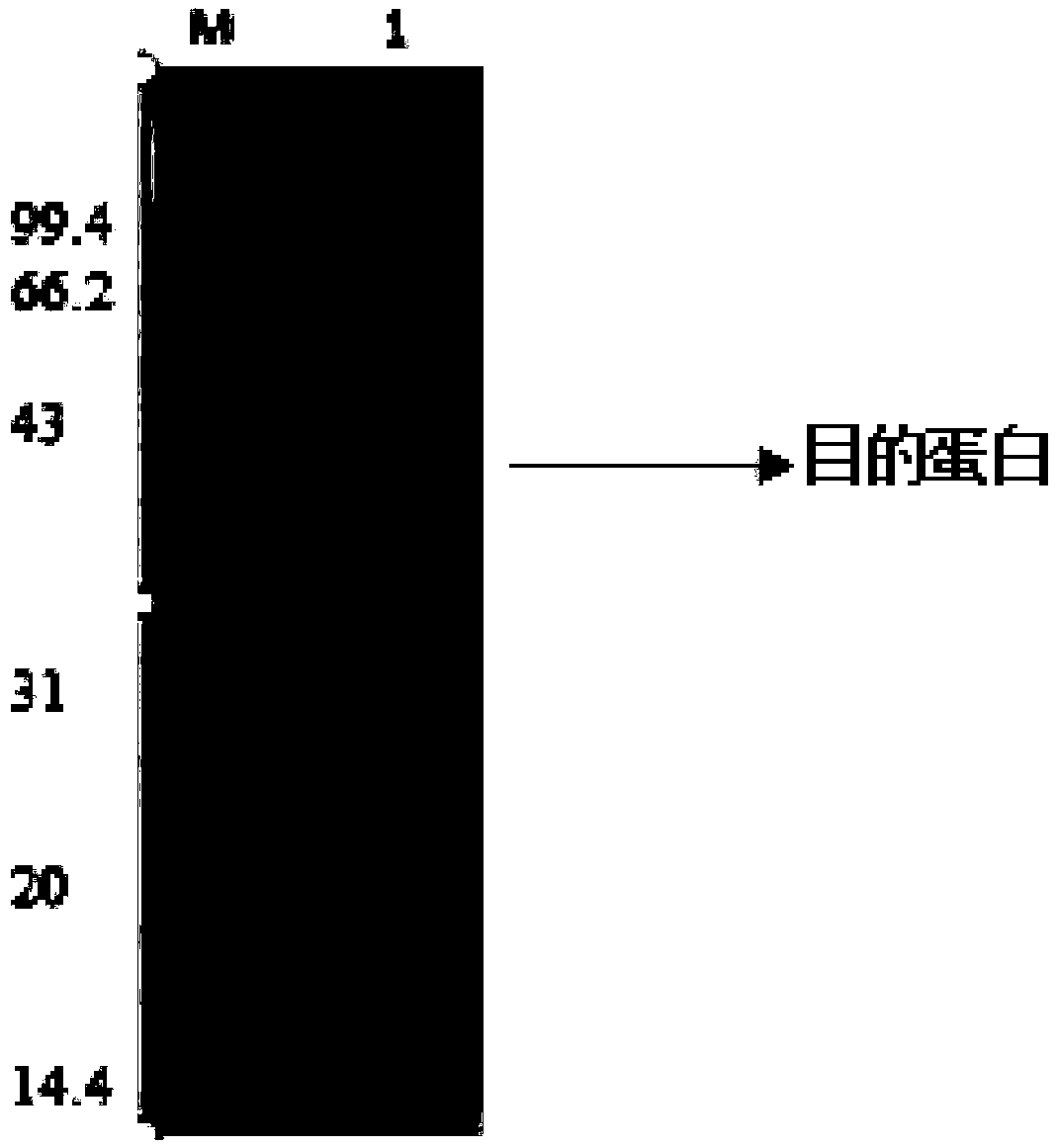
[0039] The recombinant Bacillus subtilis strain obtained in Example 2 was inoculated in LB medium, and after culturing at 37°C for 8 hours, it was transferred to 50mL TB fermentation medium with 5% inoculum size, and then placed at 37°C and 200rpm for constant temperature cultivation 2h, when the cell OD600 is 0.5-1.0, turn to 33°C and 200rpm to carry out shake-flask induction fermentation for 48h. After the fermentation, the fermentation broth was ultrasonically broken and then centrifuged, and the supernatant was the recombinant CE. The measured recombinase activity can reach 8.0U / mL, and the protein electrophoresis results show that there is a band consistent with the theoretical molecular weight at 43kDa ( figure 1 ).
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap