PIWI protein induced by nucleotide fragment and provided with specific endonuclease activity
A nuclease and fragment technology, applied in the field of PIWI protein, can solve problems such as the lack of endonuclease activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0040] Example 2: Cutting of PIWI Proteins to Related Plasmids
[0041] 1. PET28PIWI plasmid extraction
[0042] Plasmid extraction was carried out using Sangon Bioengineering Co., Ltd. Column Plasmid DNA Mini-Extraction Kit to extract the plasmids of PET28PIWI and pUC57. The two plasmids have a homologous region of about 800bp. The extracted plasmids were analyzed for quality and quantified by agarose gel electrophoresis.
[0043] 2. Cutting of related plasmids by PIWI nuclease
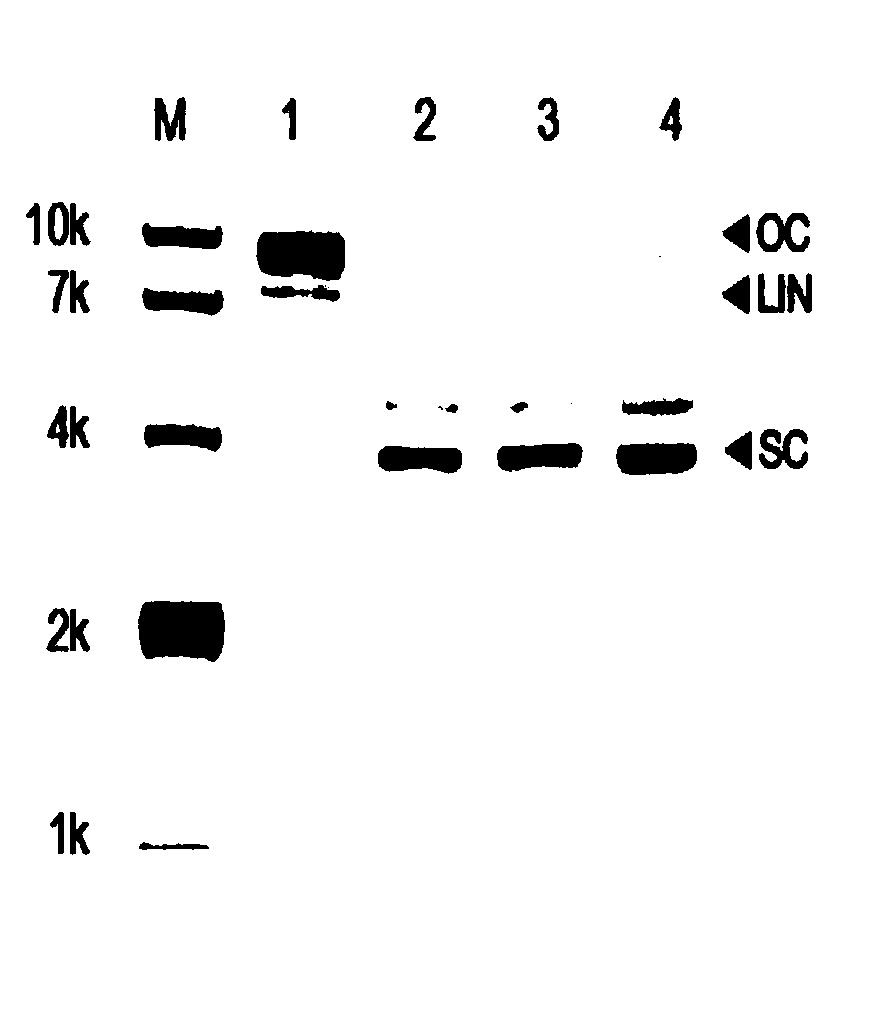
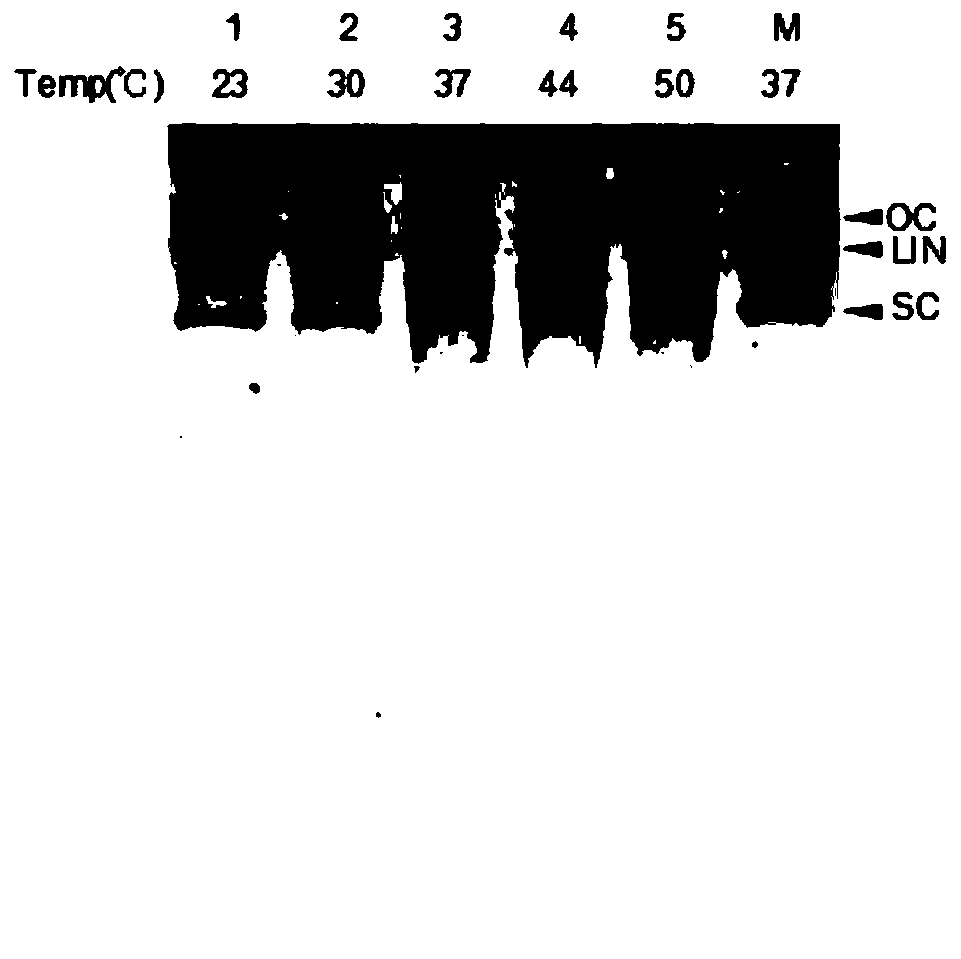
[0044] After the relevant plasmid is transferred into the host cell, a large number of small DNA / RNA fragments complementary to the plasmid itself will be produced, and the expressed PIWI protein can bind to these small DNA / RNA fragments and produce a shearing effect on the plasmid itself. The PIWI protein was mixed with the plasmid (the plasmid expressing the protein PET28-PIWI) at a molar ratio of 10:1, and reacted at 37°C for 4 hours, and the shearing efficiency was analyzed. When the molar rat...
Embodiment 3
[0045] Example 3: Analysis of PIWI-bound short nucleotide fragments and the guiding effect of 5' phosphorylated DNA fragments on PIWI nuclease directional cleavage
[0046] 1. Type analysis of short nucleotide fragments combined with PIWI
[0047] Treat PIWI with proteinase K, and degrade PIWI completely at 55°C for 2 hours; extract the above degradation products with phenol chloroform, and purify the small nucleotide fragments combined with PIWI; use RNase or DNase to treat the small nucleotide fragments respectively Processing was performed to determine whether PIWI bound DNA or RNA. The results showed that no small nucleotide fragments appeared in the electrophoresis of samples added with RNase, but small nucleotide fragments appeared in the electrophoresis of samples added with DNase, indicating that PIWI binds small RNAs in vivo ( Figure 4 ).
[0048] 2. Design of 5' phosphorylated DNA fragments
[0049] According to the sequence of the pEGFP plasmid, design a 5' phos...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com