Application of an insect detoxifying enzyme protein in degrading organophosphorus pesticides
A technology for detoxifying enzymes and pesticides, which is applied to the application field of insect glutathione-S transferase protein in the degradation of malathion, and can solve problems such as the inability to remove malathion pesticide residues and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0011] Example 1: Obtaining the LmGSTpcF1 gene of migratory locust
[0012] Using the cDNA of the resistant strain migratory locust as a template, use primers: FF 5'-3': ATGCCAGTCACCCTC, and FR 5'-3': CTACGGAGCCAGATG to clone the full-length LmGSTpcF1 gene. The system is: 2×Taq PCR MasterMix (25 μL), DNA template (1 μL), primers (1 μL each), ddH 2 O (21 μL), a total of 50 μL. After the product was recovered, purified and sequenced, SEQ ID NO:1 was obtained.
Embodiment 2
[0013] Example 2: Expression and purification of LmGSTpcF1
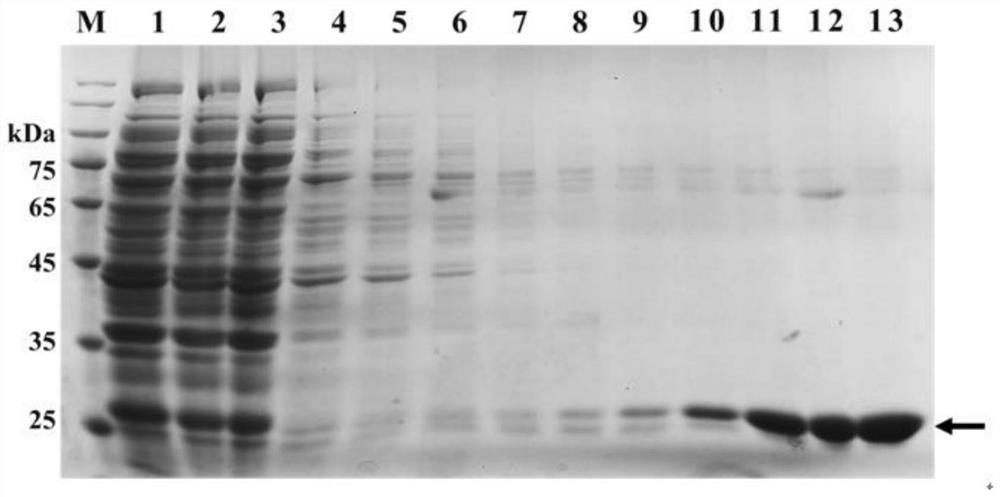
[0014] According to SEQ ID NO: 1, primers including BamHI and HindIII restriction sites were designed, WF5'-3': TAGGATCCATGCCAGTCACCCTC and WR5'-3': ATAAGCTTCTACGGAGCCAGATG were used for recombinant plasmid construction. After PCR, the product was digested with BamHI and HindIII, and recovered by 1.5% agarose gel electrophoresis. The prokaryotic expression vector pET28a was also processed synchronously. After the recovered products were mixed, buffer and T4 ligase were added, ligated overnight at 4°C, and transformed into Escherichia coli DH5α competent cells. Select the strains with accurate verification results, expand the culture, extract the recombinant plasmid, and transform it into Escherichia coli BL21 (DE3) competent cells. Pick positive colonies, after PCR verification, expand culture. Positive monoclonal colonies were inoculated into liquid LB medium (containing 50 μg·mL -1 Kanamycin) test tubes, cultiv...
Embodiment 3
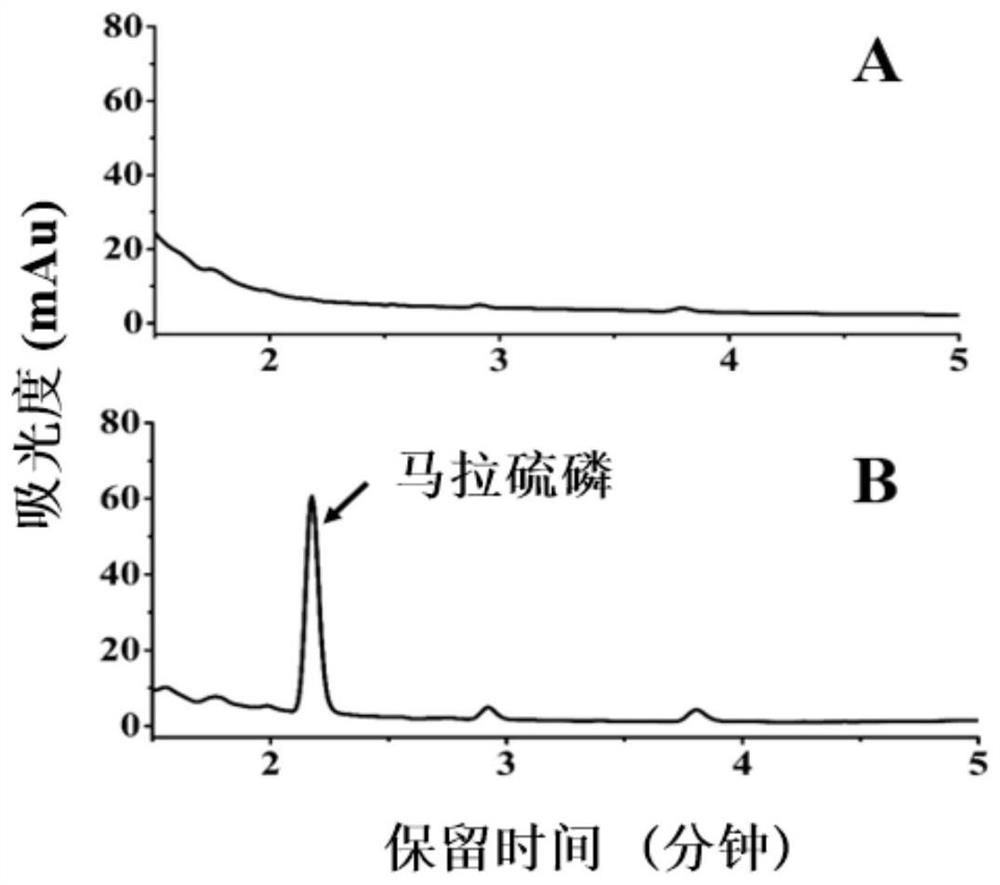
[0016] Example 3 Metabolism detection of malathion by LmGSTpcF1
[0017] The peak area of pesticides detected by HPLC was used to calculate the degree of metabolism of malathion by LmGSTpcF1. The chromatographic detection condition is that mobile phase A is acetonitrile, and B is 0.1% formic acid water. The elution gradient is A:B=80:20 isocratic elution for 8 minutes; the detection wavelength is 210nm; the column temperature is 35°C; the sample volume is 2uL; the flow rate is 0.2mL / min.
[0018] The test samples were divided into two groups, A and B. Group A was the normal response group (LmGSTpcF1+GSH+malathion); group B was the protein-free control group (PBS+GSH+malathion) with PBS buffer instead of protein. The 200uL reaction solution includes: 20uL 1.6mg / mL LmGSTpcF protein (32ug); 15uL 20mg / mL malathion (300ug), 10uL 50mM GSH (2.5mM), PBS to make up the volume to 200uL. After mixing the reaction solution, place it at 27°C, 1200r / min, and incubate for 3h; after the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com