Phosphotransketolase with increased activity and its application in the production of metabolites
A technology of phosphoketolase and metabolites, applied in the field of genetic engineering, can solve the problems of low F/XPK enzyme activity and limited application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0123] Example 1. Construction of a strain that knocks out the pfk gene, i.e. strain Z188△pfk
[0124] Corynebacterium glutamicum Z188, namely Corynebacterium glutamicum Z188. The complete genome of Corynebacterium glutamicum Z188 can be found in GenBank accession number: NZ_AKXP00000000.1 (https: / / www.ncbi.nlm.nih.gov / nuccore / NZ_AKXP00000000). The pfk gene is the 6-phosphofructokinase gene. In the genomic DNA of Corynebacterium glutamicum Z188, the nucleotide sequences of the coding frame of the pfk gene and the 1000bp parts upstream and downstream thereof are shown in sequence 2 of the sequence listing (in the sequence 2 of the sequence listing, 1001-2041 nucleotides is a coding frame, encoding the protein shown in Sequence 1 of the Sequence Listing).
[0125] Δpfk-F1: CCTC GAATTC GGATGCTGCCAATGGAATGGTGCCCAGTG;
[0126] Δpfk-R1: CTTCAAGGTT AAATTCATTGCTGGCTGTGC;
[0127] Δpfk-F2: CAATGAATTT AACCTTGAAGGAAGTTCCATTC;
[0128] Δpfk-R2: TCTA CTGCAG GGAATGATGACACCGATGGTGT...
Embodiment 2
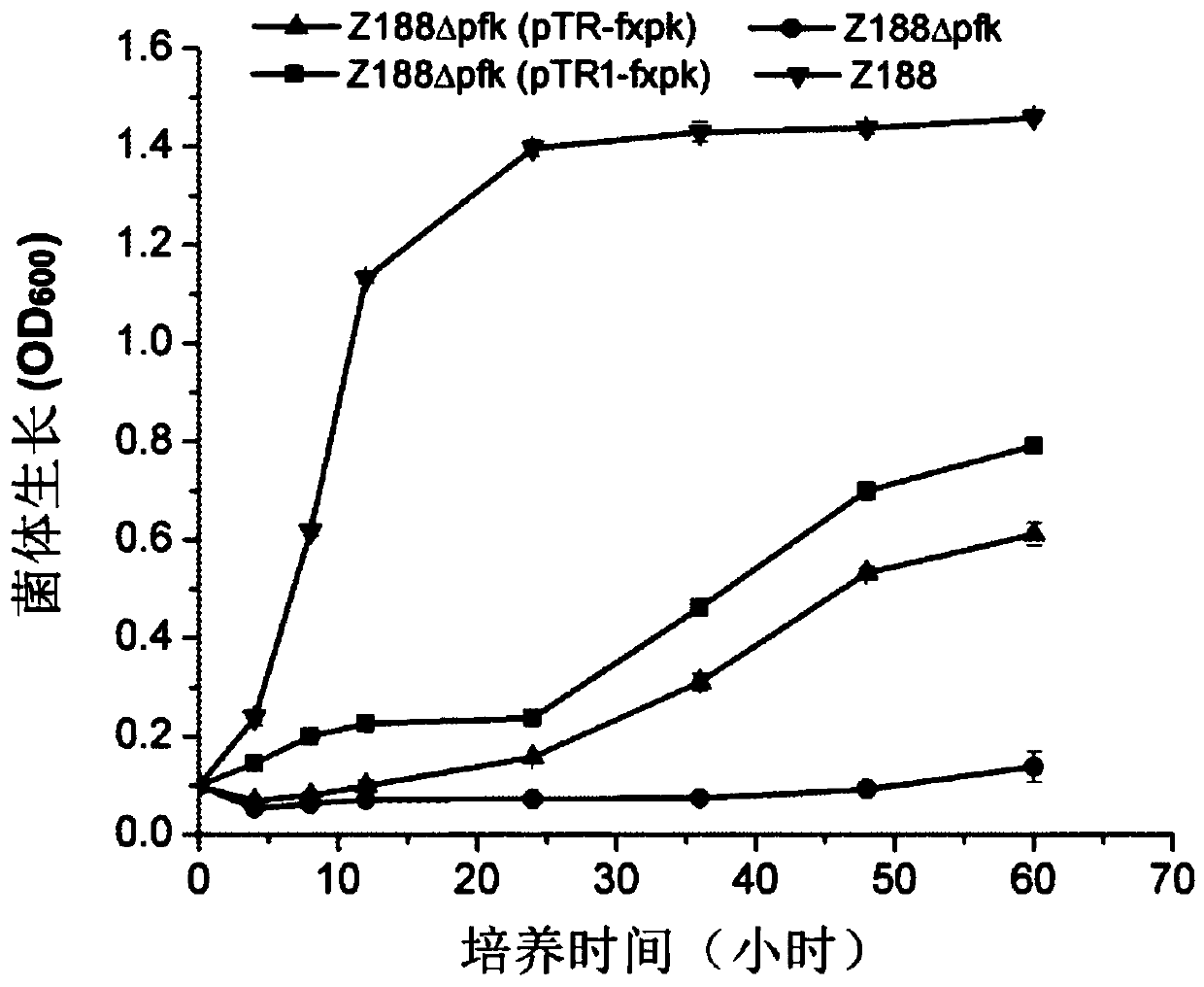
[0137] Embodiment 2, preparation and growth performance of recombinant bacteria
[0138] The phosphoketolase derived from Bifidobacterium adolescentis is shown in sequence 3 of the sequence list. The complete codon optimization is carried out on the gene encoding the protein shown in sequence 3 of the sequence listing, and the optimized gene is shown in sequence 4 of the sequence listing. The phosphoketolase shown in sequence 3 of the sequence listing is also called FXPK protein or F / XPK protein. The gene encoding FXPK protein is also called fxpk gene.
[0139] 1. Preparation of recombinant plasmids
[0140] 1. Synthesize the double-stranded DNA molecule shown in sequence 4 of the sequence table; use the synthesized double-stranded DNA molecule as a template, perform PCR amplification with a primer pair composed of F1 and R1, and recover the PCR amplification product.
[0141] F1: CTAC GAATTC GAAGGAGATATACATATG;
[0142] R1: TCAG GGATCC TCATTCGTTGTCACCCGCGGTC.
[0143...
Embodiment 3
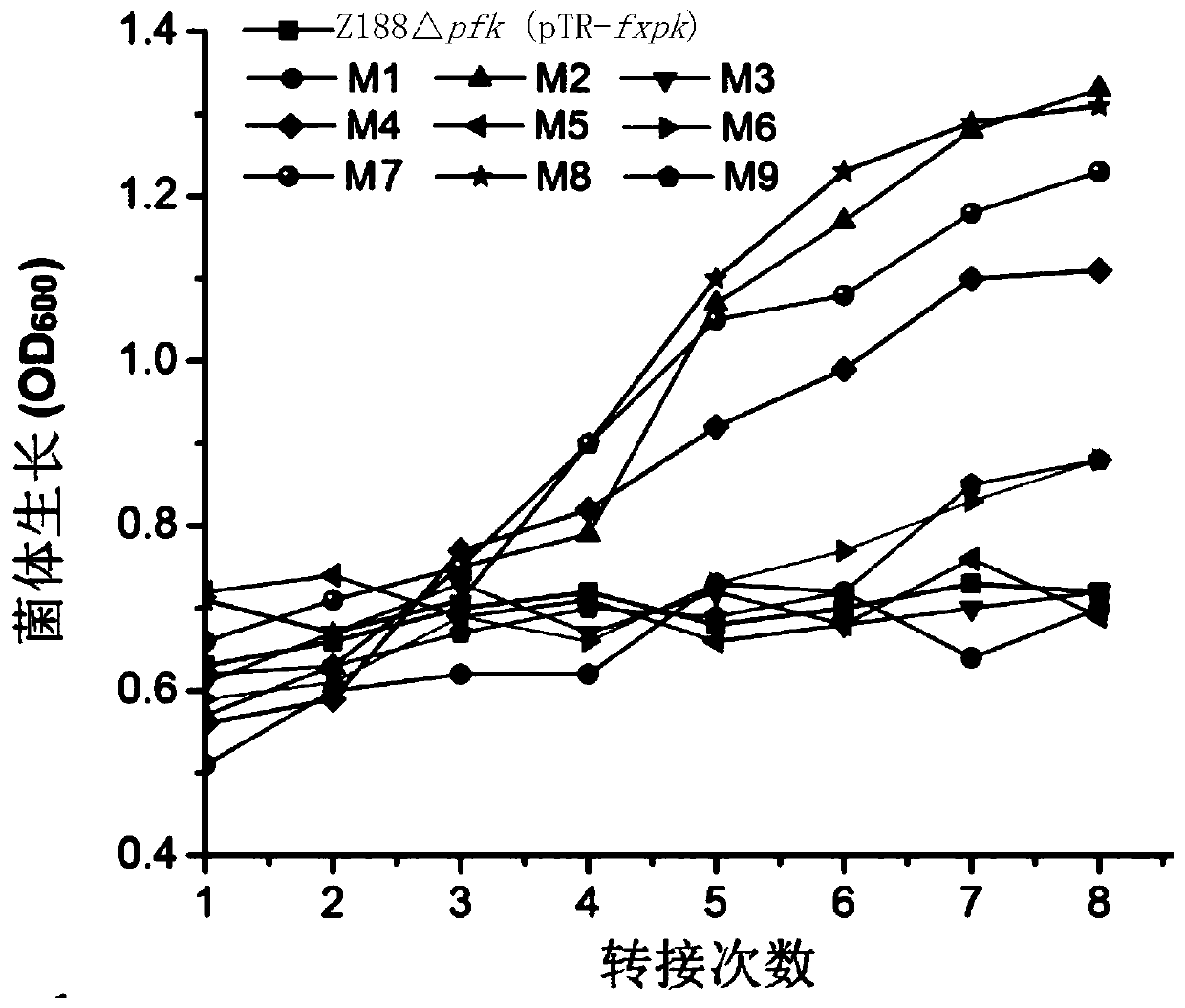
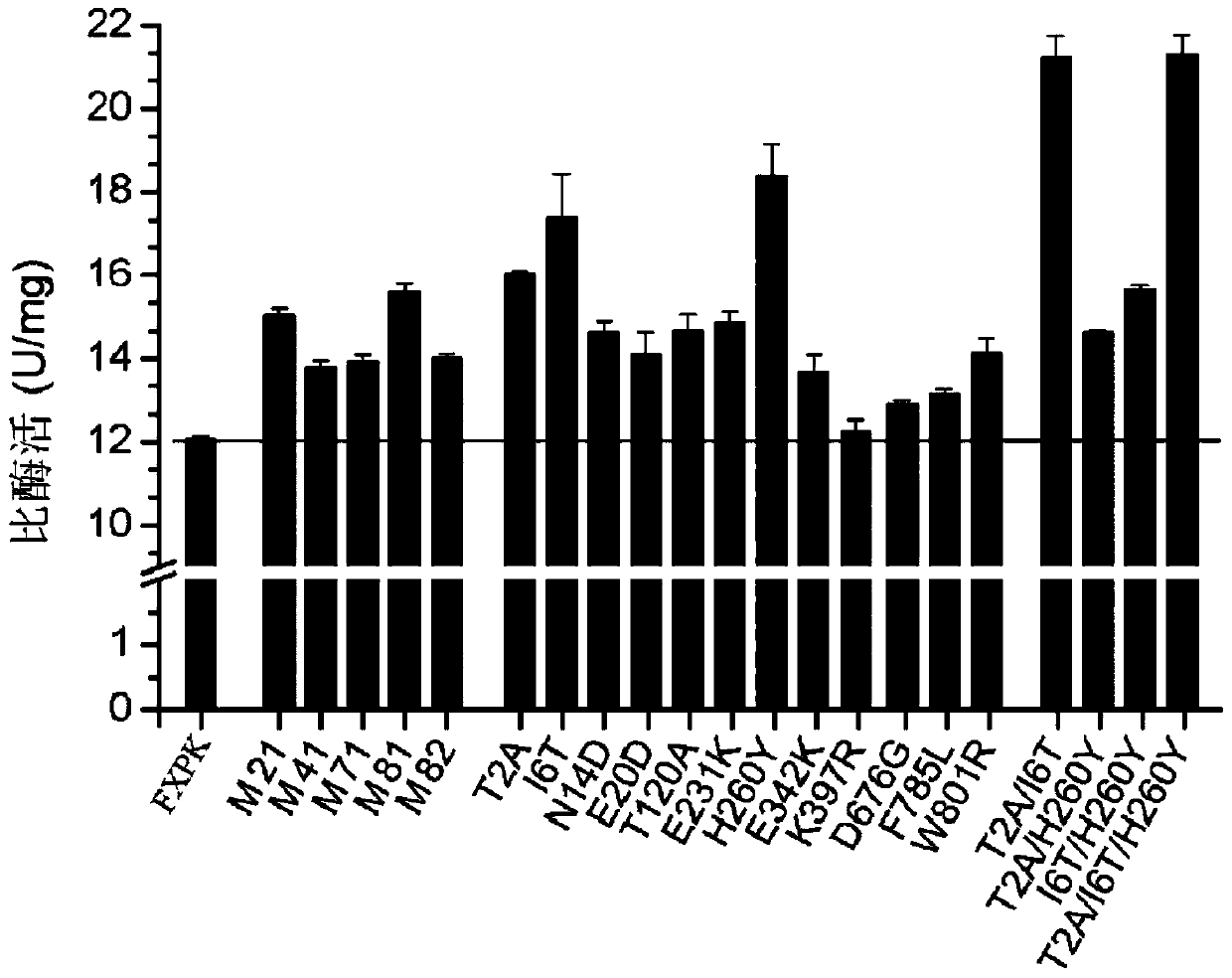
[0157] Embodiment 3, obtain the mutein that enzyme activity improves
[0158] 1. Construction of the fxpk gene mutant library
[0159] 1. Using the recombinant plasmid pTR-fxpk as a template, the primer pair composed of F1 and R1 is used for error-prone PCR amplification. Error-prone PCR amplification, using DNA polymerase (Beijing Quanshijin Biology). Three reaction systems were set up, respectively containing 0.2mM, 0.5mM or 0.8mM manganese chloride. After completing the error-prone PCR amplification, the three reactions were combined.
[0160] 2. Take the product of step 1, perform double digestion with restriction endonucleases EcoRI and BamHI, and recover the digested product.
[0161] 3. Take the recombinant plasmid pTR-fxpk, perform double digestion with restriction endonucleases EcoRI and BamHI, and recover the vector skeleton.
[0162] 4. Ligate the digested product of step 2 with the vector backbone of step 3 to obtain a recombinant plasmid.
[0163] 5. Introd...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com