SNP molecular marker combination for identifying species of puffers
A technology of molecular markers and species identification, which is applied in the determination/inspection of microorganisms, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of hybrid toxicity risks, large differences in economic value, human danger, etc. The typing operation is simple and fast, the typing accuracy is high, and the benefit effect is guaranteed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] One of the 90 fugus from different populations was arbitrarily selected as a species identification object for species identification. The specific identification steps include:
[0049] Step 1, obtaining the genome DNA of the puffer from the fin rays, muscles, or blood of a puffer to be evaluated;
[0050] Step 2: Utilize the puffer genome DNA obtained in step 1 to construct a genome-wide SNP microarray chip, and obtain the genotypes of multiple SNP molecular marker sites of puffer puffer.
[0051] Step 3, using bioinformatics software to analyze the genotypes of the multiple SNP molecular marker sites of Puffer puffer to be evaluated according to the Mendelian genetic law, analyze and obtain the typing information of Puffer puffer to be evaluated, and determine the Identification of the species affiliation of puffer puffer.
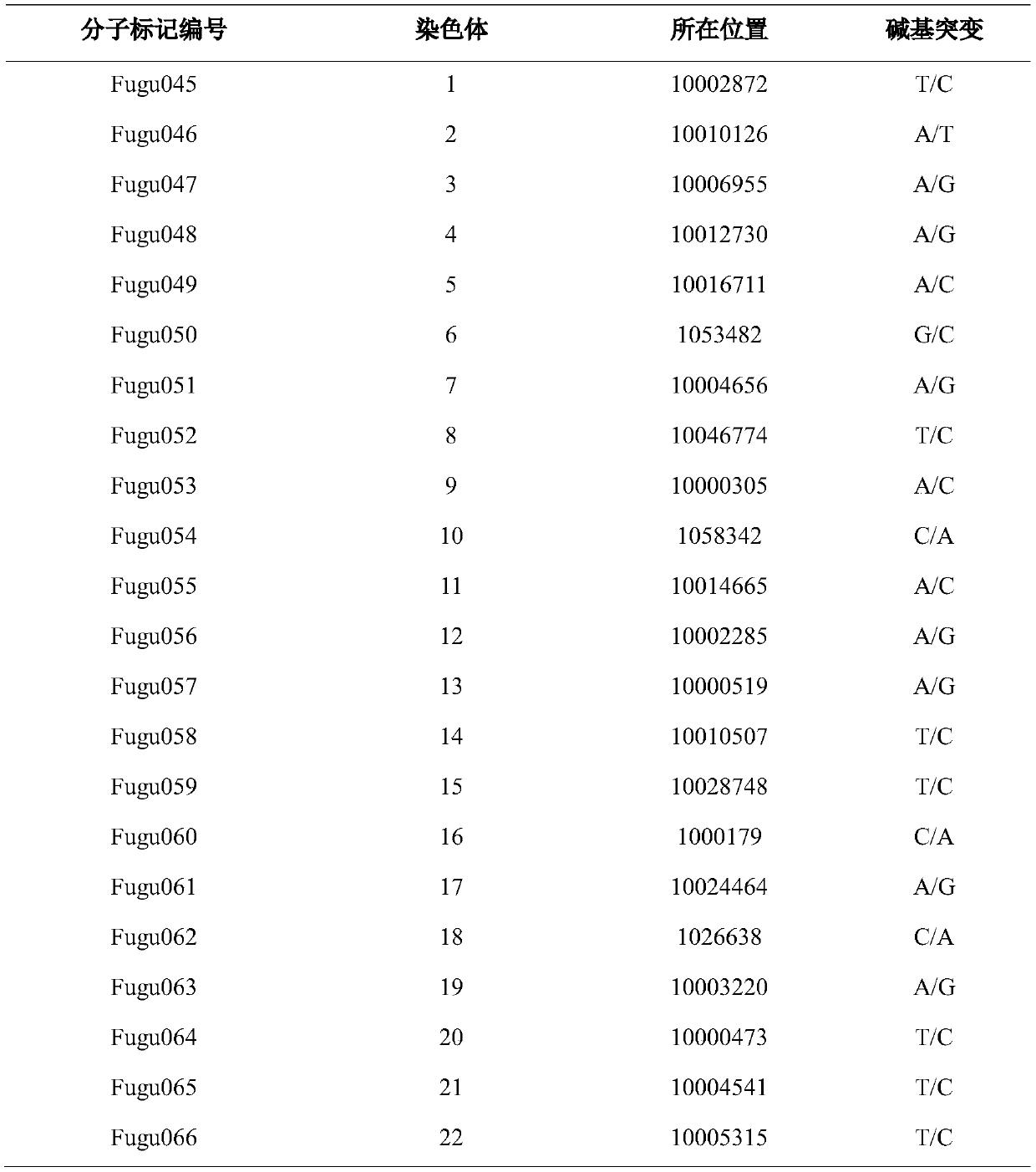
[0052] If the genotypes of the corresponding SNP loci are completely consistent with the genotype information listed in Fugu001-Fugu022, it can b...
Embodiment 2
[0058] Randomly select one of the 90 puffer fishes from different populations as the seed identification object to carry out puffer puffer seed identification. The specific identification steps include:
[0059] Step 1, obtaining the genome DNA of the puffer fish from the fin rays, muscles, or blood of a fugu fry to be evaluated;
[0060] Step 2: Utilize the puffer genome DNA obtained in step 1 to construct a genome-wide SNP microarray chip, and obtain the genotypes of multiple SNP molecular marker sites of puffer puffer.
[0061] Step 3, using bioinformatics software to analyze the genotypes of multiple SNP molecular marker sites of puffer puffer fry to be evaluated in step 2 according to the Mendelian genetic law, analyze the typing information of puffer puffer fry to be evaluated, and judge The species affiliation of puffer puffer species to be identified.
[0062] If the genotypes of the corresponding SNP loci are completely consistent with the genotype information listed...
Embodiment 3
[0068] Species identification of the parents of hybrid puffer puffer, the specific identification steps include:
[0069] Step 1, obtaining the genome DNA of the puffer from the fin rays, muscles, or blood of a puffer to be evaluated;
[0070] Step 2, constructing a whole-genome SNP microarray chip using the puffer genome DNA obtained in step 1, and obtaining the genotypes of multiple SNP molecular marker sites of the puffer puffer;
[0071] Step 3. Use bioinformatics software to analyze the genotypes of multiple SNP molecular marker sites of Pufferfish to be evaluated according to the Mendelian genetic law, and analyze the typing information of Pufferfish to be evaluated, such as its corresponding SNP The genotype of the site partly matches any of the genotype information listed in Fugu001~Fugu022 or Fugu023~Fugu044 or Fugu045~Fugu066 or Fugu067~Fugu088 or Fugu089~Fugu110, and partly matches Fugu001~Fugu022 or Fugu023~Fugu044 or Fugu045 ~Fugu066 or Fugu067~Fugu088 or Fugu089...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com