A method for designing a CRISPR-induced RNA library
A design method and library technology, applied in genomics, instrumentation, sequence analysis, etc., can solve the problems of impracticality and high computational cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] The present invention will be further described below in conjunction with examples.
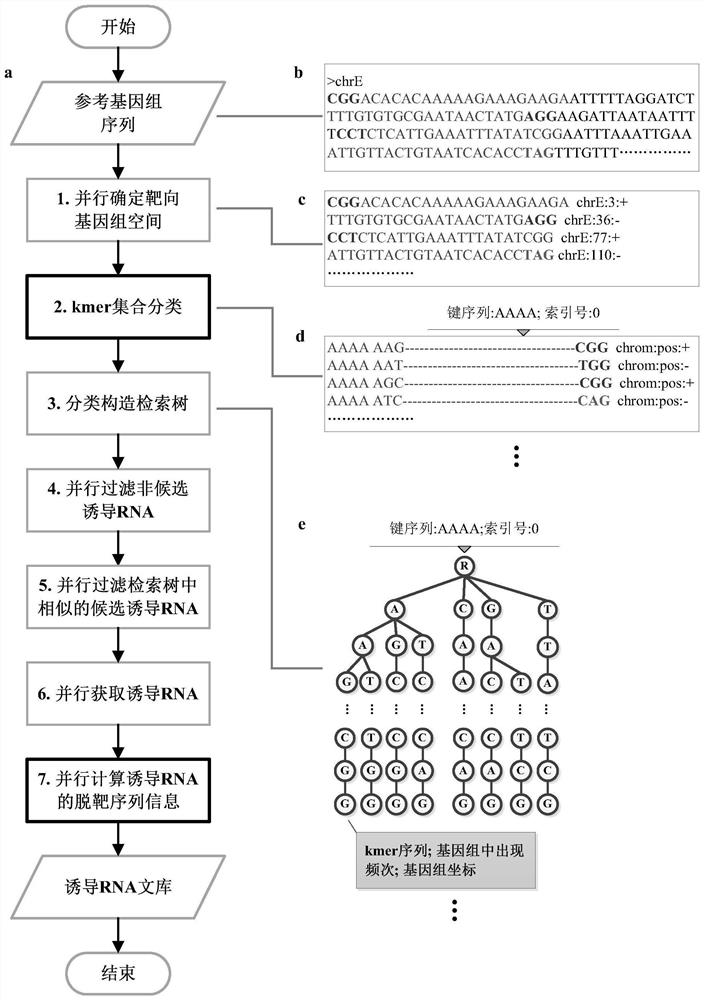
[0039] This embodiment provides a method for designing a CRISPR-induced RNA library, which specifically includes the following steps:
[0040] Step 1: Determine the targetable genomic space in parallel.
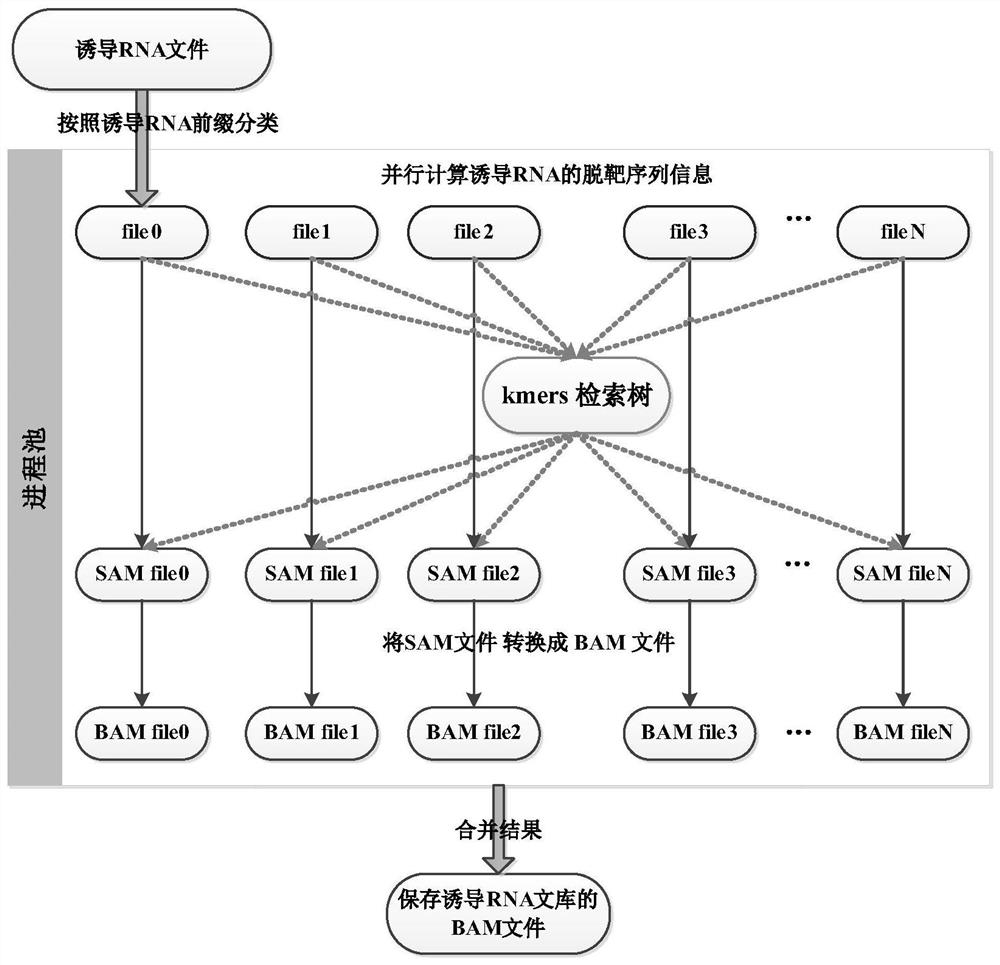
[0041] In order to generate the induced RNA library desired by the user, the method also allows the user to input the reference genome FASTA file, the length of the induced RNA, the standard PAM (protospacer adjacent motif), the PAM position relative to the induced RNA target sequence, non- Standard PAM, and Hamming distances M and Q. Such as figure 1 As shown in (b, c), based on the parameters given by the user, the algorithm scans the kmer prefixed or suffixed by standard PAM and non-standard PAM in the reference genome, as well as related information, such as its corresponding coordinates and directions (used to record kmer in position and encoding direction in a reference seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com