A Knockout Mutant Strain of Streptococcus suis type 4 galr gene sh1510δgalr
A technology of Streptococcus suis and gene knockout, applied in the field of genetic engineering, can solve the problem that SS4 virulence factor has not been reported, and achieve the effect of simple and efficient construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Construction of Mutant SH1510ΔGalR
[0026] 1) Primers
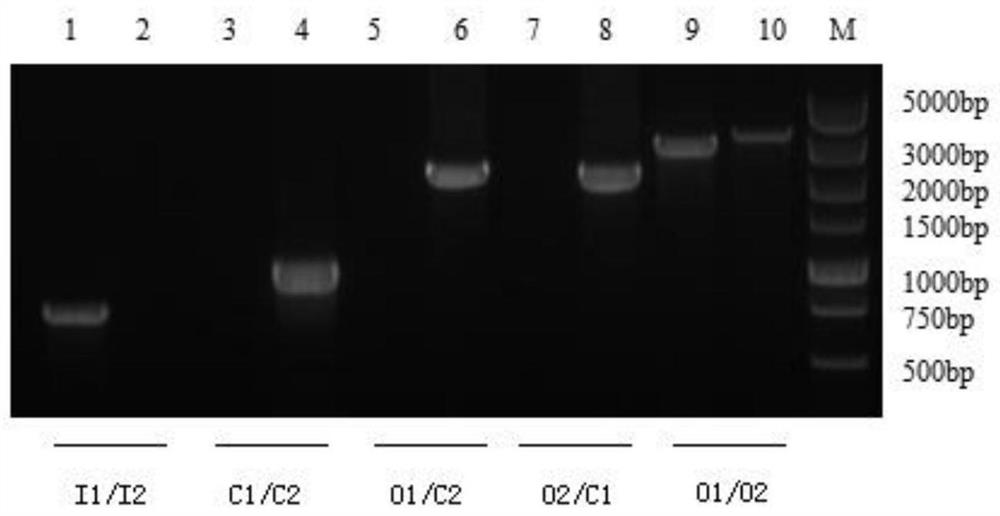
[0027] The details of the primer sequences designed in this experiment are listed in Table 1. L1 / L2, R1 / R2 respectively amplify the gene sequence of the upstream and downstream homology arms of the GalR coding gene; C1 / C2 amplifies the chloramphenicol resistance gene; I1 / I2 amplifies the ORF internal gene sequence of the GalR gene (for deletion strains); design a pair of primers O1 / O2 outside the upper and lower homology arms of the GalR gene, and amplify the total length of a gene sequence including the GalR gene, the upper and lower homology arms genes and the outside (for external detection) .
[0028] Table 1 Primer sequence list
[0029]
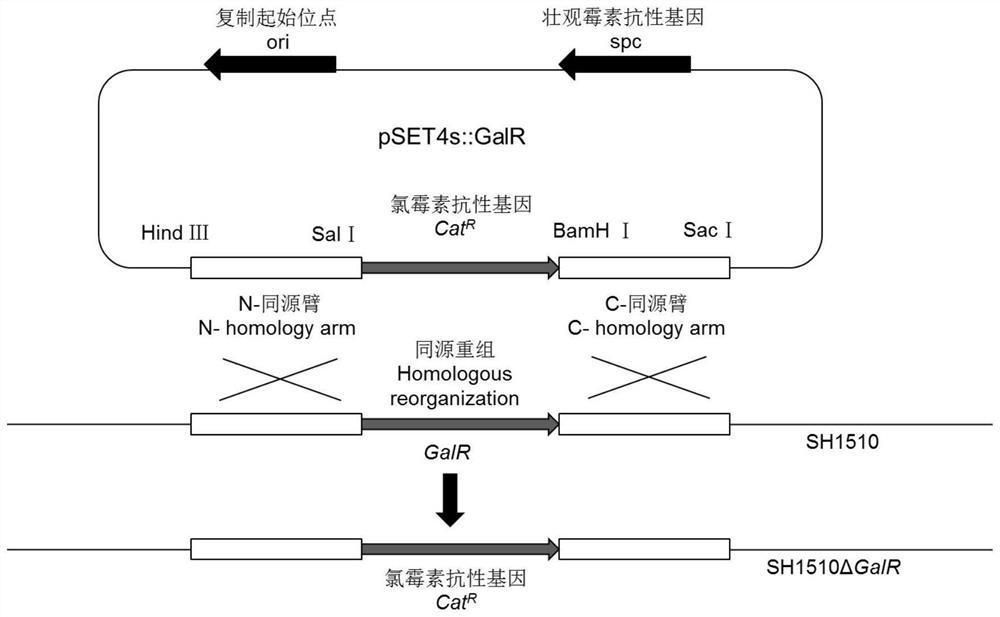
[0030] 2) Construction of the deletion strain SH1510ΔGalR
[0031] A. Construction of the gene deletion plasmid pSET4s::GalR as follows figure 1 As shown, primers L1 / L2, R1 / R2 and SH1510 DNA templates were used to amplify the upstream and downstream homolo...
Embodiment 2
[0042] Example 2 Gram staining
[0043] The cryopreserved parent strain SH1510 and deletion strain SH1510ΔGalR plates were streaked for recovery, and single colonies of the parent strain and deletion strain were picked and inoculated in THB broth, 200 r / min, shaking culture at 37°C until logarithmic growth phase, The parental strain SH1510 and the deletion strain SH1510ΔGalR were subjected to smear, Gram staining and microscopic examination, and the morphology of the two strains was observed and compared under the oil lens.
[0044] exist Figure 4 It can be seen that the two strains are round or oval, arranged in chains, and have similar lengths, without significant difference.
Embodiment 3
[0045] The comparison of different sugar utilization rates of embodiment 3
[0046] The basal medium was prepared according to Tang Yulong's method, and six bottles of basal medium were prepared for the test. Two bottles were used as a group, and 10mmol / L glucose, sucrose and D-galactose were added respectively. Resuscitate the bacteria, pick a single colony of the parental strain SH1510 and the deletion strain SH1510ΔGalR and inoculate them in THB broth, 200r / min, shake at 37°C until OD 600 =0.7, centrifuge at 12000r / min for 10min, discard the supernatant, wash the bacteria twice with sterilized physiological saline, and resuspend the bacteria in an equal volume. Add the bacterium solution of the parental strain and the deletion strain to the medium of the three groups at a ratio of 1%, and measure the OD every 1 h 600 value until bacterial OD 600 The value can only be stopped when it reaches a stable value, the test is repeated 3 times, and the hourly OD is calculated 600...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com