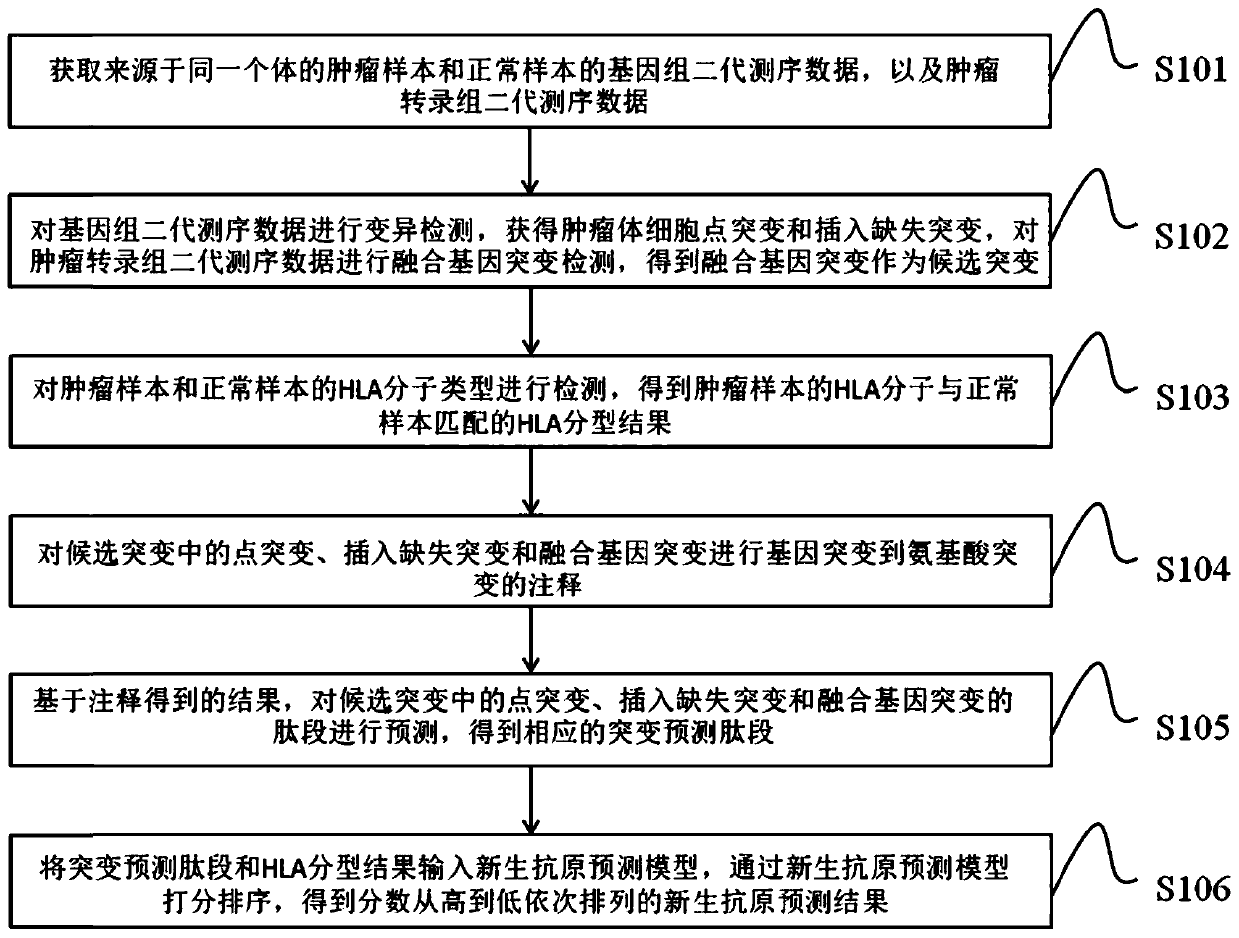
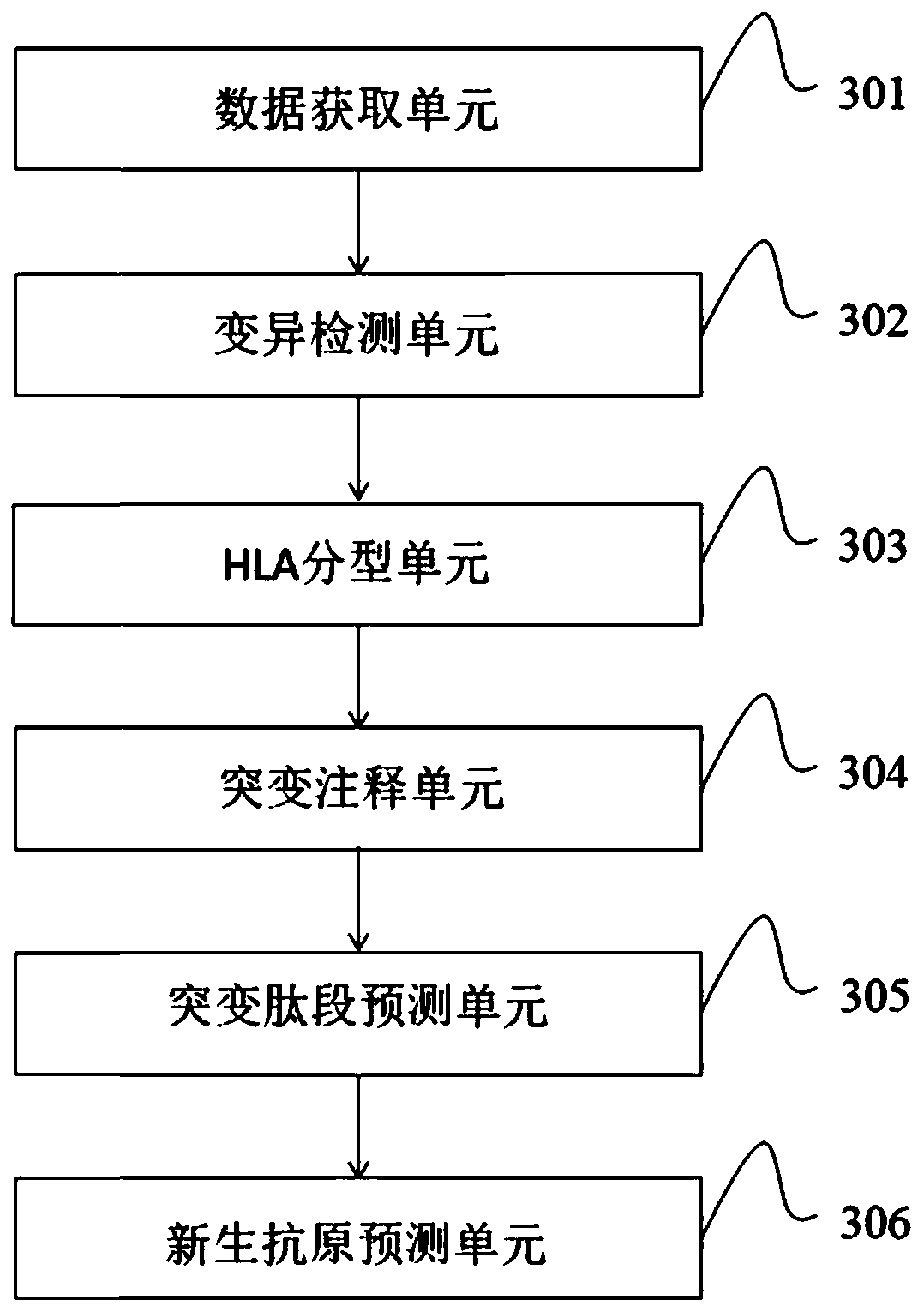
Neoantigen prediction method and device based on next-generation sequencing and storage medium
A new generation of next-generation sequencing technology, applied in the field of bioinformatics, can solve the problem of whether the mutant peptides are presented on the cell surface, etc., and achieve convenient treatment, high sensitivity and specificity, and direct prediction results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1: Model training
[0081] 1. Data preparation
[0082] Get the positive training data from the neoantigen peptide mass spectrum database, take the peptides between 6-16 amino acids, and randomly intercept the peptides larger than 11 amino acids to the length between 9-11 amino acids, these data are mass spectra Validation of neoantigen data presented to the surface of tumor cells. Using the data in the SwissProt protein database, randomly intercept peptides with a length between 9-11 amino acids, and participate in training as a negative data set. The HLA-I typing information corresponding to the neoantigen peptide was obtained from the database. The negative / positive peptides are one-hot coded, the HLA-I typing information is one-hot coded, and input into the model.
[0083] 2. Model training module
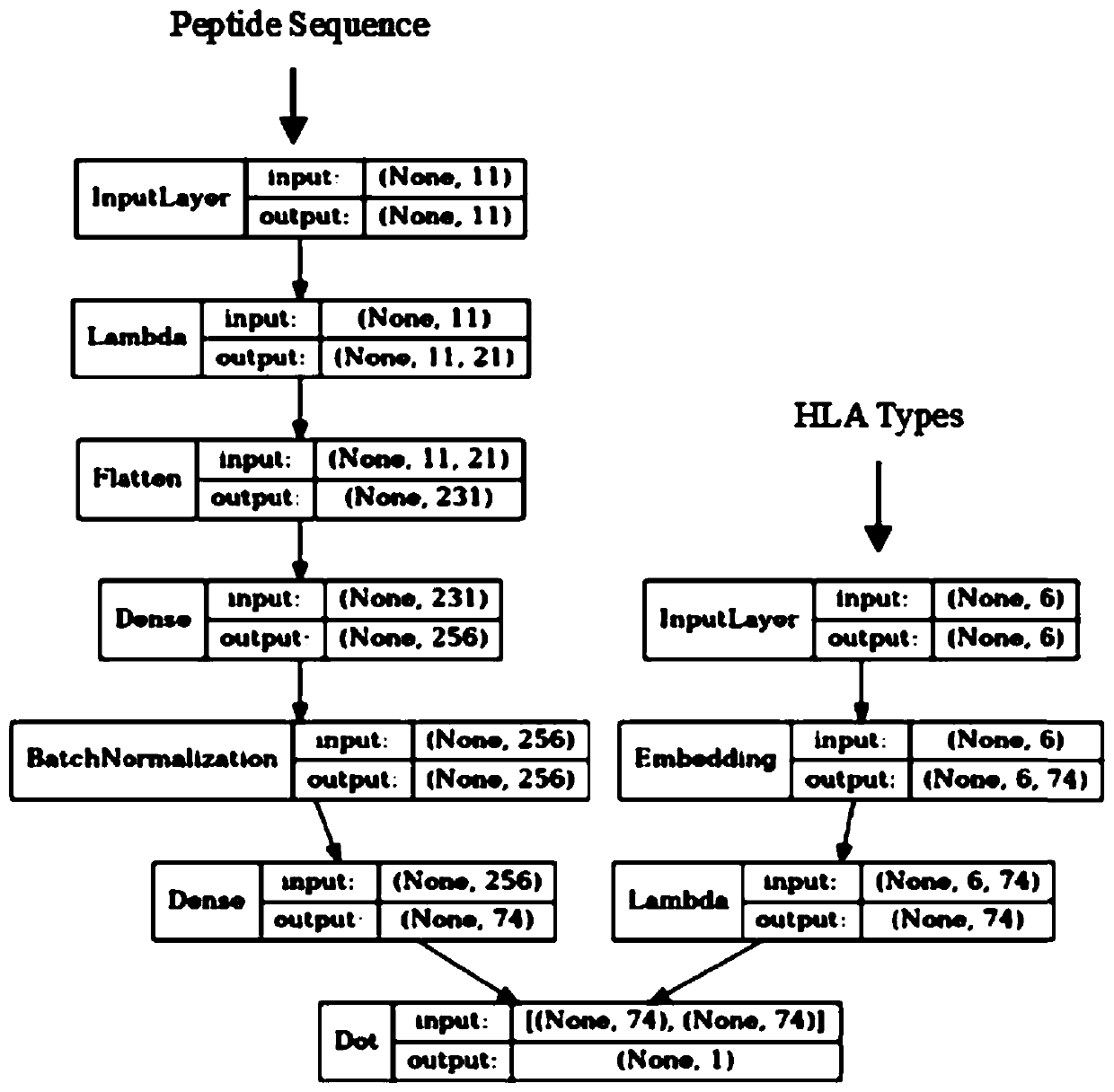
[0084]Model training uses the above prepared data, and the model includes the following components: (1.1) Input Layer (input layer), if the input peptide is ...
Embodiment 2
[0087] Example 2: Neoantigen prediction
[0088] In this embodiment, the samples used are provided by TESLA (Tumor Neoantigen Screening Alliance), and the neoantigen peptides that can bind to the corresponding HLA are verified through experiments. The principle of experimental verification is based on tetramer technology, check the reaction of pMHC (peptide / MHC conjugate) with T cells, and obtain positive / negative peptides.
[0089] The five samples are numbered 1, 2, 10, 103, and 210. The specific steps of sample detection in this example are as follows: obtain the HLA-I typing of each sample; obtain all mutant peptides of each sample, and intercept Include mutated amino acids to 11 amino acids in length, if there are no 11 amino acids, use X to fill in; input the trained neoantigen prediction model; sort the output according to the neoantigen score, and take the peptide with a score greater than 0.1 as a positive neoantigen result.
[0090] As a control, the published soft...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com