Process method for combined analysis of single-cell scRNA-seq and scATAC-seq
A scRNA-seq, combined analysis technology, applied in sequence analysis, instrumentation, genomics and other directions, to achieve the effect of scientific and reasonable structure, simplified operation process, safe and convenient use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
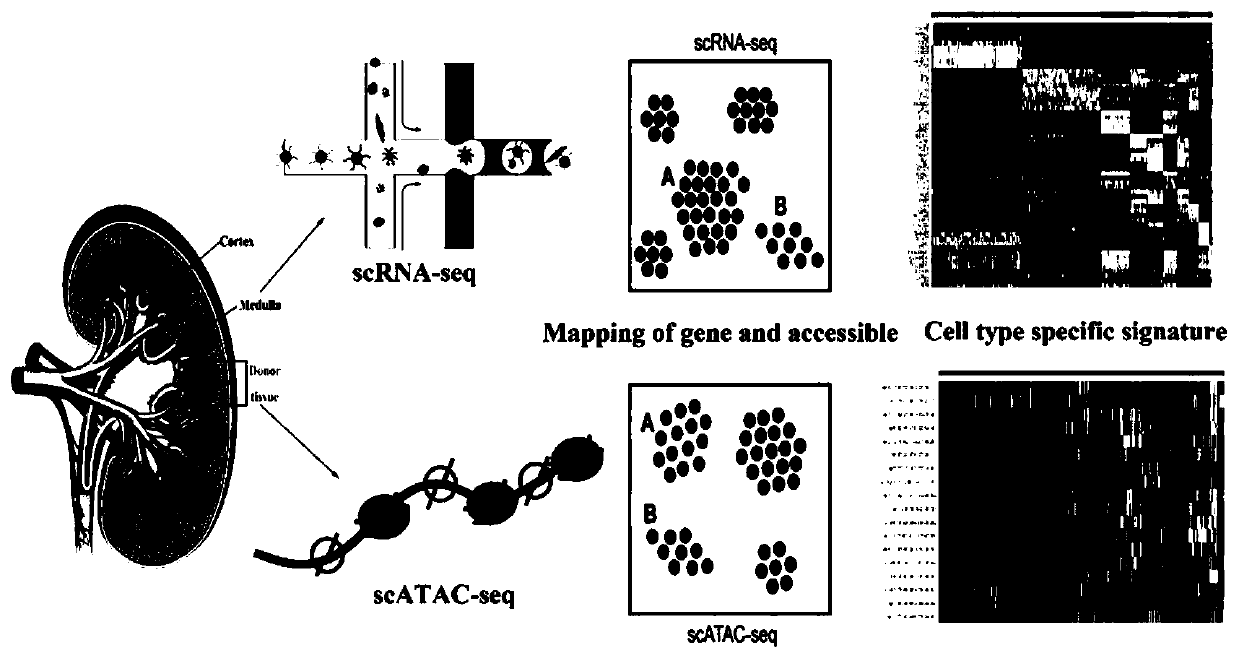
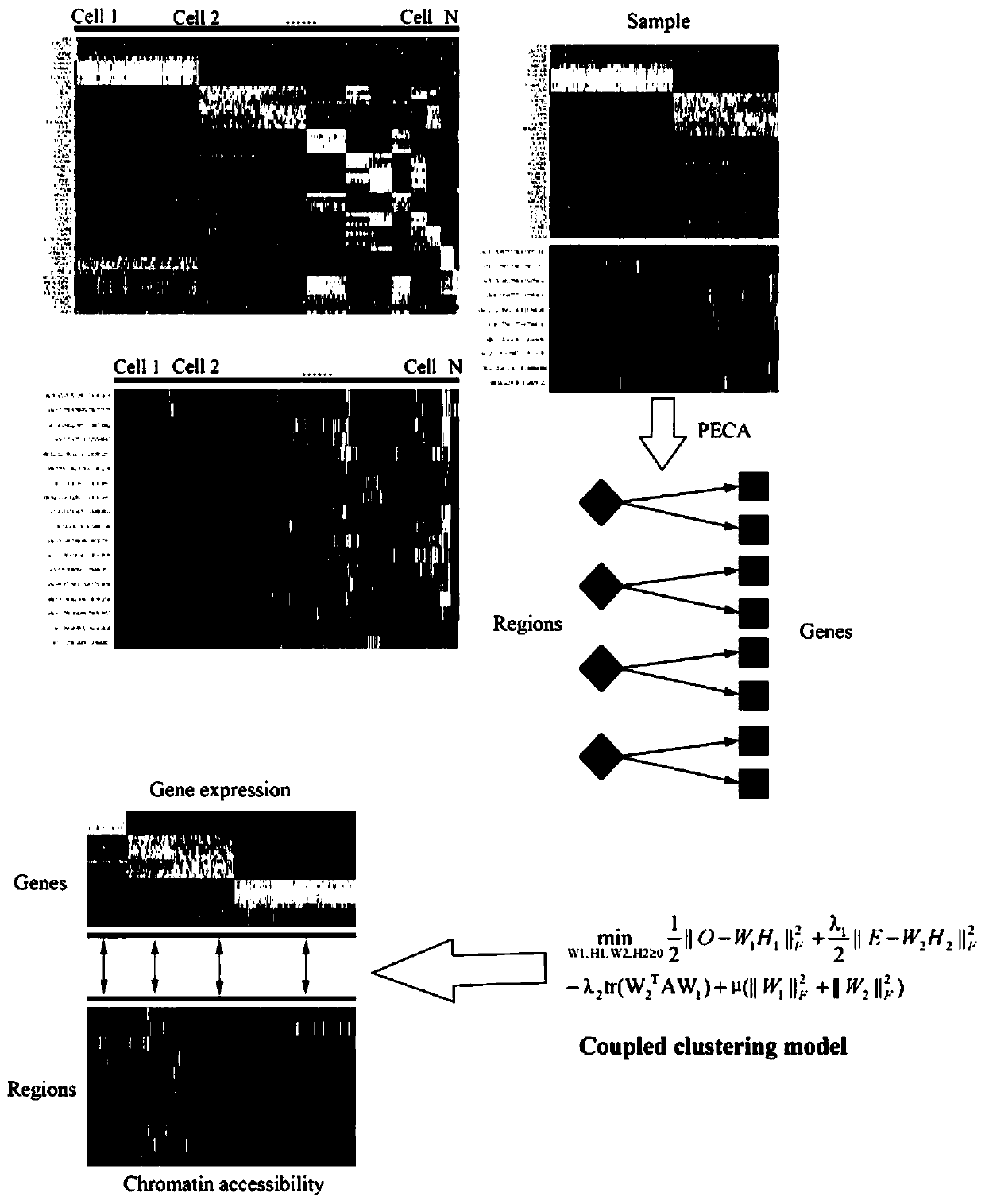
[0031] Example: such as figure 1 As shown, the present invention provides a technical solution, a process method for joint analysis of single-cell scRNA-seq and scATAC-seq, including scRNA-seq analysis, scATAC-seq analysis, scRNA-seq and scATAC-seq joint analysis;
[0032] The scRNA-seq analysis comprises the following steps:
[0033] A1. Raw data processing;
[0034] A2. Difference analysis and cell clustering;
[0035] A3, TF search;
[0036] The scATAC-seq analysis comprises the following steps:
[0037] B1. Raw data processing;
[0038] B2. Search for the position and intensity of the signal peak;
[0039] B3. Correlation analysis and difference analysis;
[0040] B4. Search for transcription factors.
[0041] According to the above technical scheme, the step A1 uses the Fastq format file of the raw data obtained by sequencing as the input file to perform raw data processing, and uses the sequence of the fastq file to run the data using the parameter "cellranger cou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com