Primer suitable for sequencing DNA-coding compound sequencing library
A technology for sequencing primers and compound libraries, which is applied in the field of primers for DNA-encoded compound sequencing libraries to construct sequencing, can solve the problems of low sequencing quality, influence authenticity, poor base diversity, etc., to improve sequencing quality, reduce detection costs, and improve The effect of data utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Establishment of a DNA-encoded compound sequencing library by conventional methods
[0038] 1. PCR amplification
[0039] According to the following amplification primer sequences and amplification methods, the screened DNA-encoded compounds were amplified, and two biological replicates were set up, labeled S1 and S2 respectively, and a negative control was set at the same time.
[0040]
[0041]
[0042] The PCR amplification system is 30 μL of screened DNA-encoded compounds, 4 μL of 5’-primers, 4 μL of 3’-primers, 100 μL of Q5 hot start ultra-fidelity 2X master mix and 62 μL of ddH 2 O, a total of 200 μL per sample.
[0043] The PCR reaction program is: first step: 98°C for 10 minutes; second step: 98°C for 10 seconds, 55°C for 10 seconds, 72°C for 10 seconds, and repeat 18 times; third step: 72°C for 10 seconds Cool to 4 °C after 1 min.
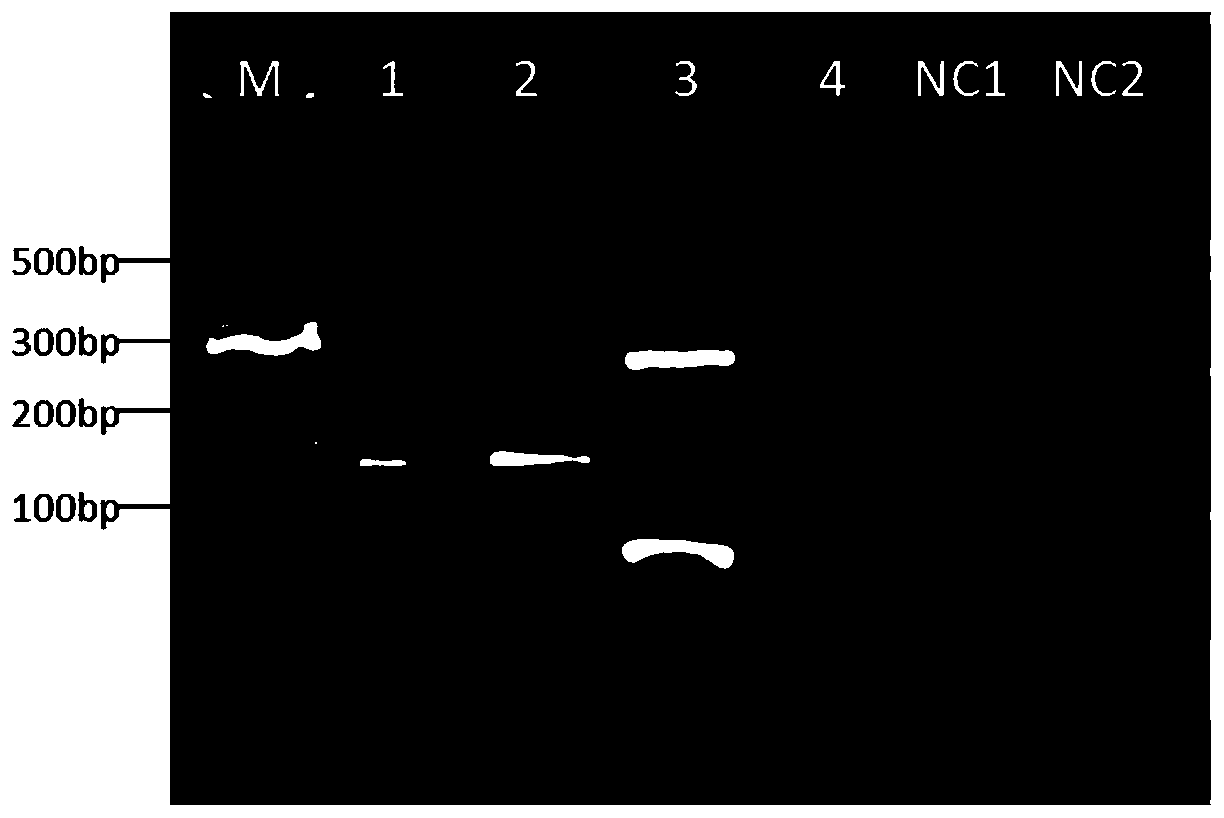
[0044] Sampling and electrophoresis to detect PCR products. Take 5 μL of each sample for agarose gel electroph...
Embodiment 2
[0056] Example 2 Establishment of a DNA-encoded compound sequencing library by the method of the present invention
[0057] According to the following amplification primer sequences and amplification methods, amplify the same screened DNA-encoded compound as in Example 1, set up two biological repeats, respectively marked as S1-ONE and S2-ONE, and set negative control.
[0058] 1. Primers:
[0059]
[0060] 2. How to build a database:
[0061] The PCR amplification system was 30 μL of screened DNA-encoded compounds, 4 μL of 5’-primers, 4 μL of 3’-primers, 100 μL of Q5 hot-start ultra-fidelity 2X master mix and 62 μL of ddHO, a total of 200 μL for each sample.
[0062] The PCR reaction program is: first step: maintain 98°C for 10 minutes; second step: maintain 98°C for 30 seconds, 70°C for 30 seconds, 72°C for 30 seconds, and repeat 18 times; third step: maintain 72°C for 10 seconds Cool to 4 °C after 1 min.
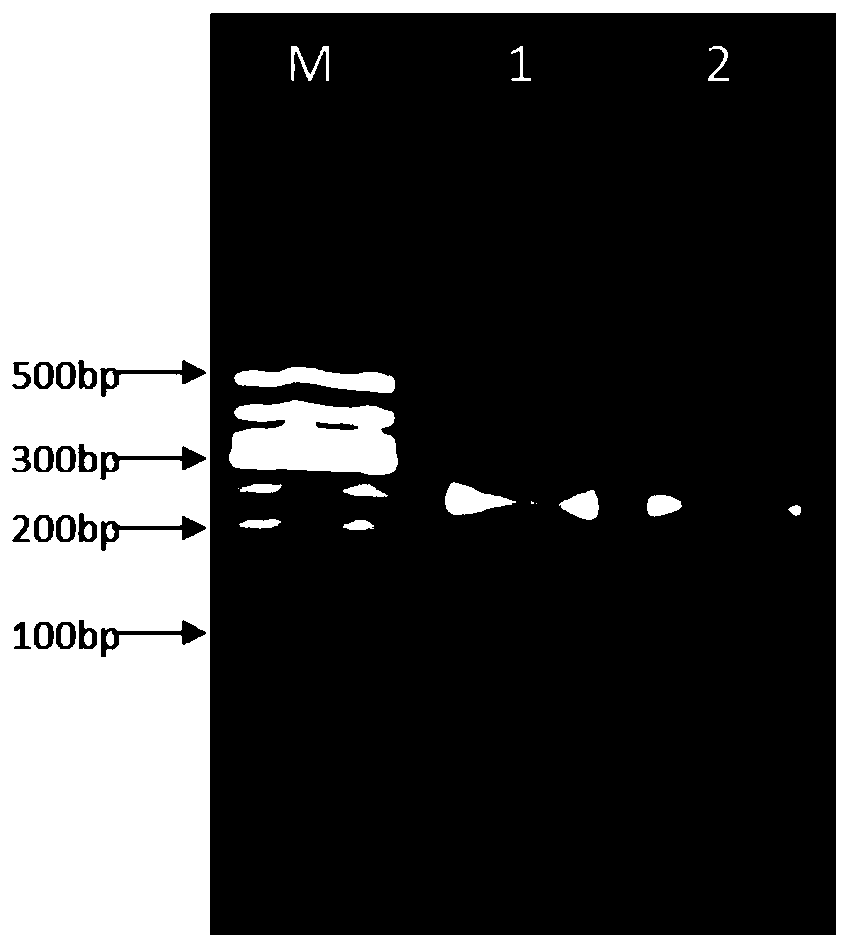
[0063] Take 5 μL of each sample for agarose gel electrophores...
Embodiment 3
[0070] Example 3 Comparison of the sequencing quality between the conventional library construction method without linker sequence and the library construction method of the present invention with linker sequence
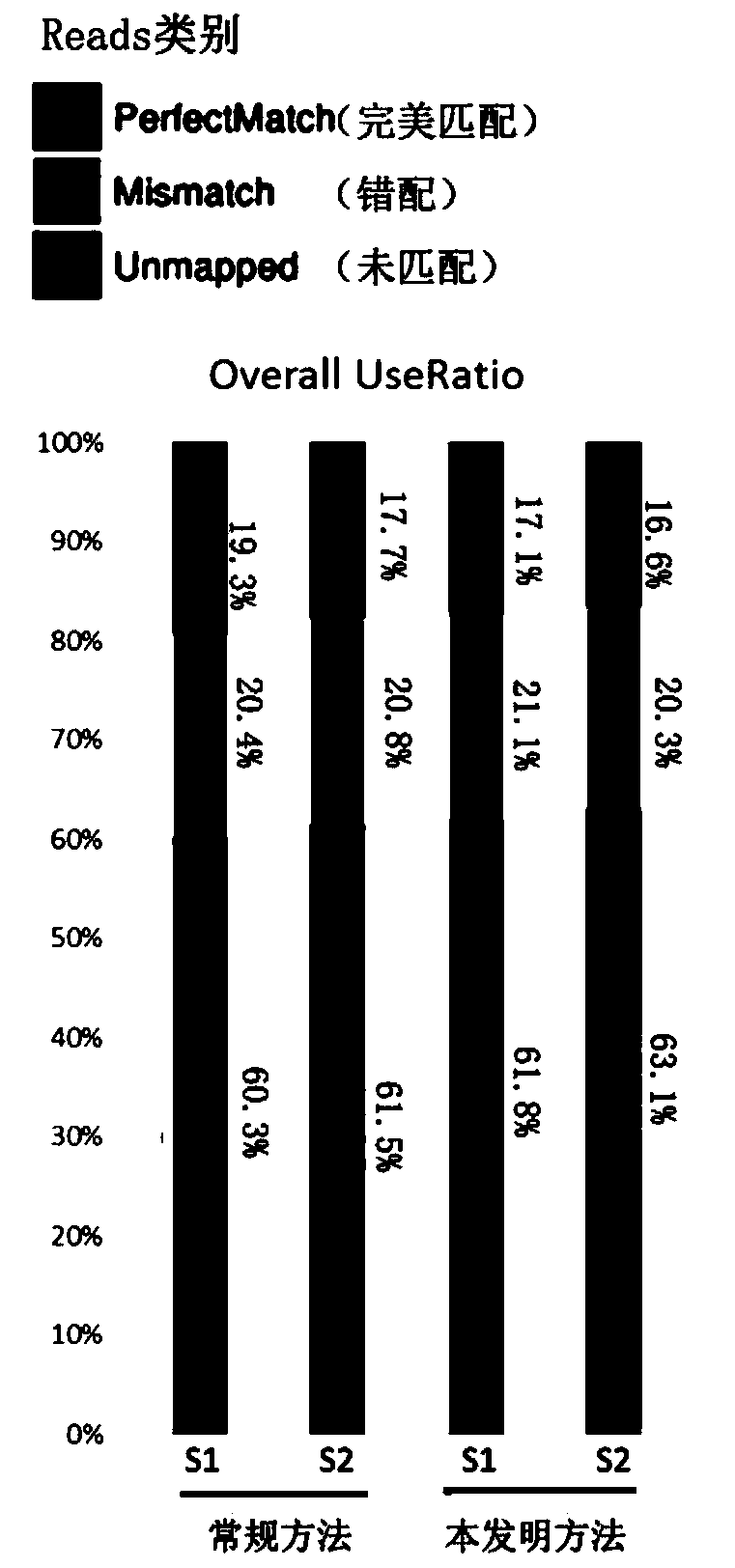
[0071] In next-generation sequencing, a corresponding quality value is given for each base measured, and this quality value is used to measure the accuracy of sequencing. The higher the quality value (Q), the smaller the probability (P) that the base is detected incorrectly, and its calculation formula is Q=-101gP. Therefore, when the Q≥30 of a single base (sometimes also written as "quality ≥ Q30"), it means that the probability of the base being detected incorrectly is less than 1‰. For the entire sequencing reads (reads), the sequencing quality is generally considered to be good when the proportion of bases with sequencing quality > Q30 is greater than 85%.
[0072] The following will compare the Q30 ratio obtained by the corresponding sequencing reaction of the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com