Method for synthesizing nucleic acid drug conjugate
A technology for drug conjugates and nucleic acid drugs, which can be used in drug combinations, pharmaceutical formulations, medical preparations with inactive ingredients, etc., and can solve problems such as increased synthesis costs, limitations, and reduced synthesis efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
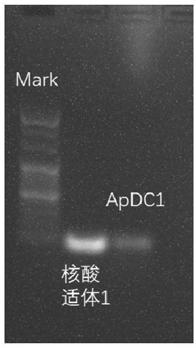
[0128] The nucleic acid aptamer drug complex constructed by the PCR method of embodiment 1
[0129] 1. Nucleic acid aptamer drug complex constructed by PCR method
[0130] The drug used in this example is 5F uracil (drug 1), also known as 5FU (is it 5FdU or 5FU). 5FdU is a base analogue, and 5FdU is connected to an upstream primer by PCR synthesis to obtain an upstream primer with 5FdU connected to its 5' end.
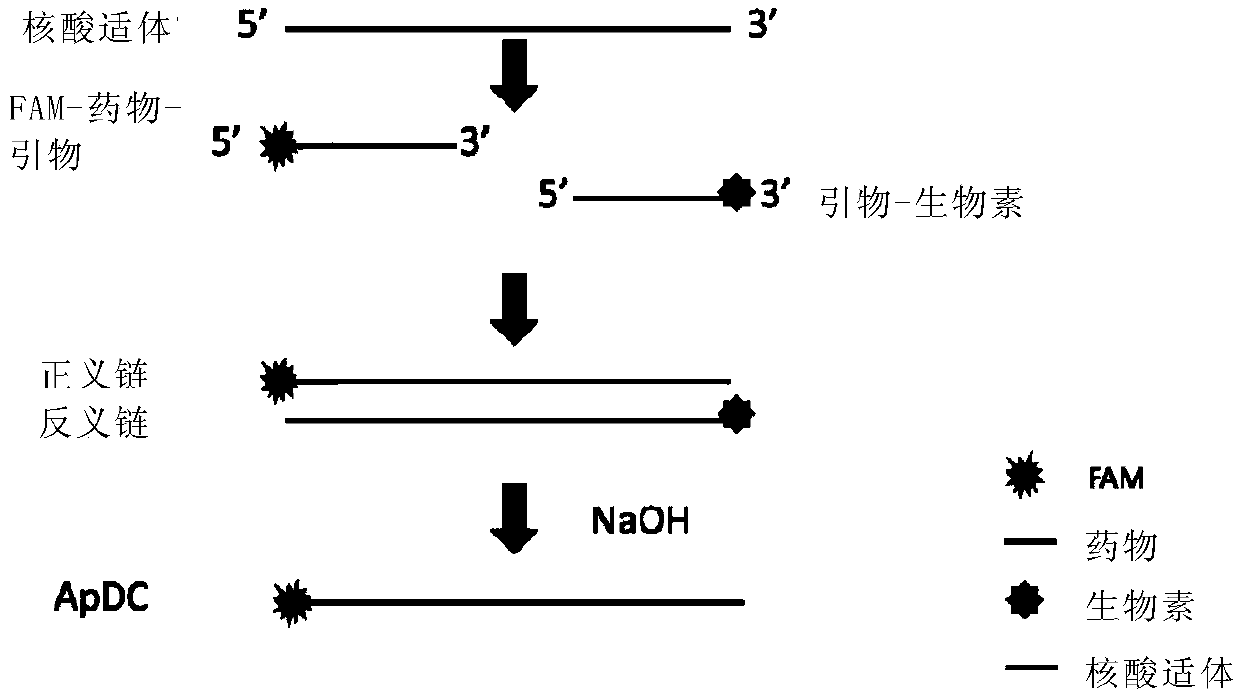
[0131] Construction of nucleic acid aptamer drug complexes (Aptamer-Drug-conjugates, ApDC) by PCR method, the process is as follows figure 1 shown. Wherein, the template is aptamer sgc8 (aptamer 1), which targets PTK7 protein, and its sequence is TTTTTTATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA (SEQ ID NO.: 3).
[0132] Upstream primer: 5'-FAM-5FdU5FdU5FdU-TTTTTTATTCTAACTGCTG-3'(SEQ ID NO.:1)
[0133] Downstream primer: 5'-Biotin-TCTAACCGTACAGTATTT-3'(SEQ ID NO.:2)
[0134] The PCR system is: 100 μL of 10X buffer, 80 μL of dNTP, 300 μL of upstream and downstream pr...
Embodiment 2
[0148] The bispecific nucleic acid aptamer-drug complex constructed in Example 2
[0149] In a similar manner to the above, two or more nucleic acid aptamer fragments are synthesized, with a few A or T bases between the nucleic acid aptamers to achieve double-specific or even multi-targeting. The drug used in this example is 5F uracil, same as Example 1. Construct nucleic acid aptamer drug complex by PCR method, the process is as follows figure 1 shown. Wherein, the template is the nucleic acid aptamer sgc8-R50, wherein sgc8 targets PTK7 protein and is highly expressed on the surface of leukemia and colon cancer cells, while R50 (aptamer 2) targets lung cancer cells, and its sequence is TTTTTTATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGATTTTTTAAAGGGCGGGGGTGGGGTGGTTGGTAGTTGTTTTTTCTGTTTC (SEQ ID NO. :5).
[0150] Upstream primer: 5'-FAM-5FdU5FdU5FdU-TTTTTTATTCTAACTGCTG-3'(SEQ ID NO.:1)
[0151] Downstream primer: 5'-Biotin-GAAACAGAAAAACAAC-3'(SEQ ID NO.:4)
[0152] The PCR meth...
Embodiment 3
[0156] The drug-containing nucleic acid aptamer drug complex constructed by the PCR method of embodiment 3
[0157] The drug of this embodiment also includes 5FdC as a base analogue, which can be connected in series with the primers simultaneously with 5FdU, and a subsequent PCR reaction is performed to synthesize ApDC with two drugs.
[0158] Specifically, the synthetic primer 5'-FAM-5FdU5FdU5FdC5FdC-TTTTTTATCTAACTGCTG-3' (SEQ ID NO.: 1), and other downstream primers can be used to connect two drugs in series on the same nucleic acid sequence. The specific implementation method is the same as embodiment column 1.
[0159] The results show that the nucleic acid aptamer drug complex ApDC coupled with two drugs, 5FdC and 5FdU, can selectively target the target well, and has good targeting specificity, less toxic and side effects, and can effectively play the role of the two drugs. effect.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com