GTP cyclohydrolase I gene folE and application
A hydrolytic enzyme, pfed760-fole technology, applied in the fields of hydrolytic enzymes, applications, genetic engineering, etc., can solve problems such as functions to be determined, and achieve the effect of improving screening efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1: GTP cyclohydrolase I gene folE knockout homology arm clone
[0058] 1. PCR amplification of upstream and downstream homology arms
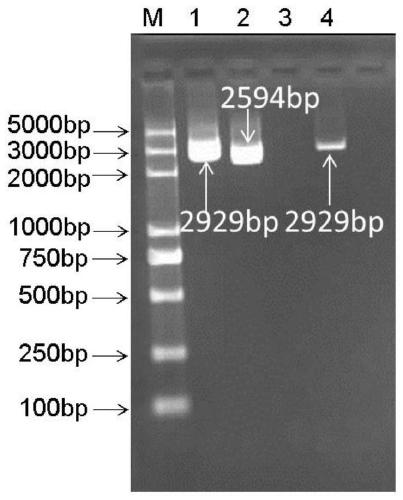
[0059] The genome of food-borne Lactobacillus plantarum YM4-3 was extracted using the Bacterial Genomic DNA Extraction Kit (Betaike Biotechnology Co., Ltd., China), and the specific operation was performed according to the kit instructions. The upstream 970bp sequence of the folE gene coding region is used as the upstream homology arm, and the downstream 1001bp sequence is used as the length of the downstream homology arm; the extracted genome is used as a template, and the primer pair is used
[0060] up-EF(5'-CCC AAGCTT GAACGGCGTAATGGTCAAC-3', the underline is the Spe I restriction site)+up-ER(5'-TTGAATTGACCACTGTAATTAATTATCAAATGGTGCGG-3') and
[0061] down-EF(5'-CCGCACCATTTGATAATTAATTACAGTGGTCAATTCAA-3')+down-ER(5'-GC ACTAGT ACACGGTCAACCGGCCTGG-3', the underline is the Hind III restriction site) to amplify the upstream ...
Embodiment 2
[0068] Embodiment 2: folE gene knockout vector construction
[0069] Use restriction endonucleases Spe I and Hind III to carry out synchronous digestion of the sequenced correct gene knockout fragment and the temperature-sensitive plasmid pFED760 respectively. The digestion system is: Spe I, 1 μL; Hind III, 1 μL; 1×H buffer , 2 μL; gene knockout fragment or pFED760, 10-16 μL; add sterilized deionized water to 20 μL, digest at 37°C for 4 hours; recover the digested product, according to the target gene:vector = 4:1-2:1 (molar ratio ) after sample addition, T4 DNA ligase was added and ligated at 16°C for 12-16 hours. The ligation product was introduced into Escherichia coli DH5α competent cells by the heat shock transformation method, and then spread on the erythromycin-LB solid plate. After culturing overnight at 28°C, extract the plasmid from 10 to 15 single colony cells, and use Spe I and Hind III enzyme digestion to obtain a positive plasmid, named pFED760-folE.
Embodiment 3
[0070] Embodiment 3: folE gene knockout strain construction
[0071] 1. The folE gene knockout vector is introduced into Lactobacillus plantarum competent cells
[0072] Lactobacillus plantarum competent cells were prepared according to the method reported by Fei Yongtao (2015, master's degree thesis of South China University of Technology). Add 10 μL gene knockout vector pFED760-folE to 90-100 μL Lactobacillus plantarum competent, mix gently, and transfer to pre-cooled electric shock cup after 5 min in ice bath, and conduct electric shock according to the parameters of 12.5kv / cm, 200Ω. After the electric shock is completed, quickly add 900 μL of fresh MRS culture solution to the electro-cup, blow and mix gently with a pipette tip, transfer the mixture to a 1.5 mL sterile centrifuge tube, and incubate at 28°C for 2.5-3 hours to recover the cells; After cultivation, the bacterial solution was centrifuged at 8,000-10,000 rpm for 3 minutes, 900 μL of the supernatant was discarde...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com