Method for assessing prognosis or risk stratification of liver cancer by using cpg methylation variation in gene
A methylation and prognostic technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1. Selection of DMPs related to the pathogenesis of liver cancer
[0081] sample
[0082] Liver cancer samples were obtained from 184 liver cancer patients at Seoul National University Hospital to screen DNA methylated regions associated with liver cancer development. For normal tissues corresponding to liver cancer tissues, a normal control group was used.
[0083] Using a column-based DNA extraction method (PureLink TM Genomic DNA Mini Kit, Invitrogen; PureLink TM Genomic DNA Mini Kit, Invitrogen) and beads (Bead) method of DNA extraction (MagListo TM 5M Genomic DNA Extraction Kit, Bioneer; MagListo TM Genomic DNA (genomic DNA) was extracted from each sample using 5M Genomic DNA Extraction Kit (Bioni). The extracted genomic DNA was quantified using a spectrophotometer (nanodrop), and the state of the DNA was electrophoresed on a 1.5% agarose gel to confirm whether it was degraded or not.
[0084] bisulfite treatment
[0085] When the genomic DNA is trea...
Embodiment 2
[0094] Example 2. Selection of Diagnostic Marker Candidates
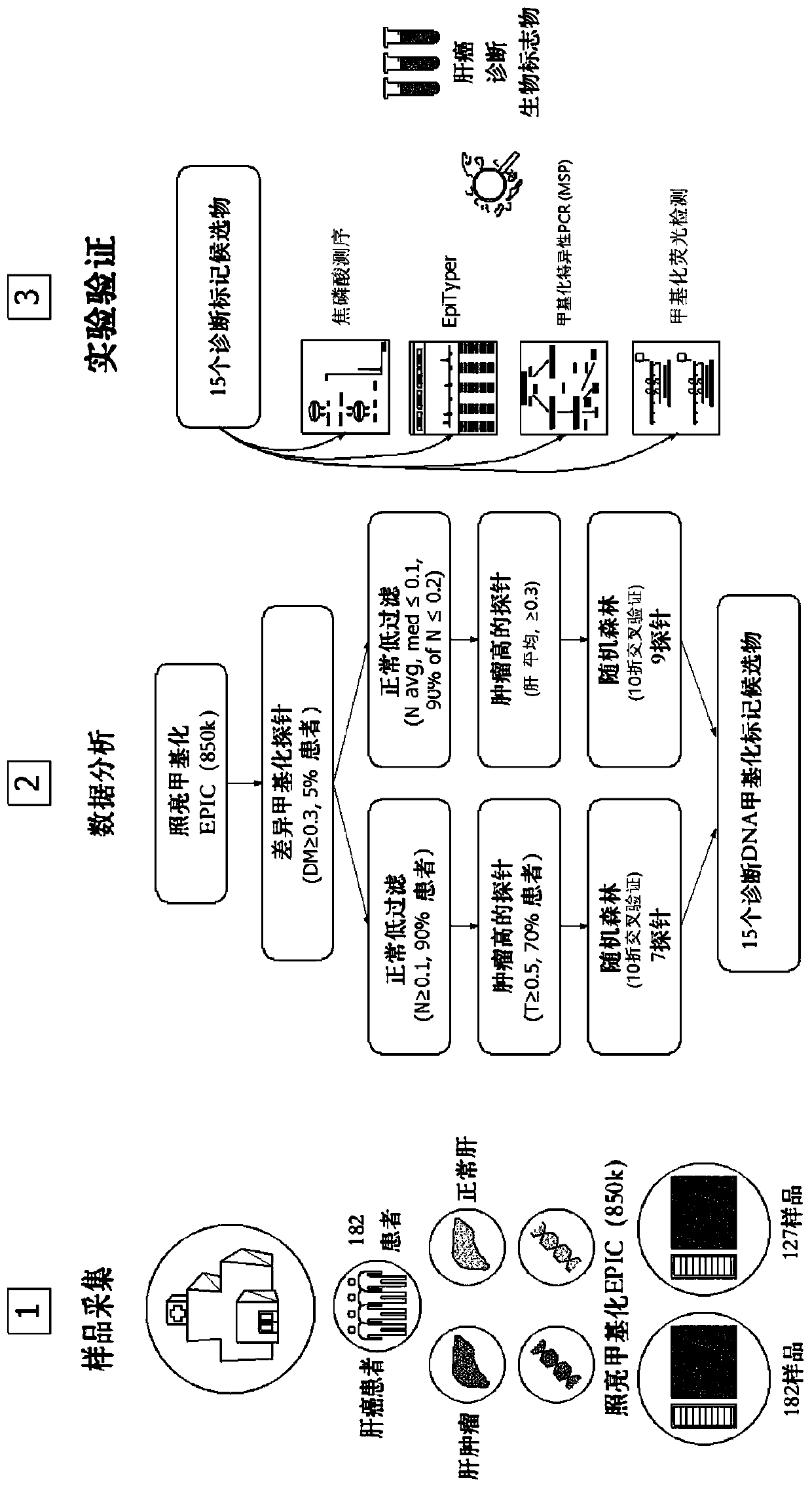
[0095] refer to figure 1 , DNA was extracted from 182 cases of liver cancer and corresponding normal liver samples, followed by InfiniumMethylation EPIC BeadChip.
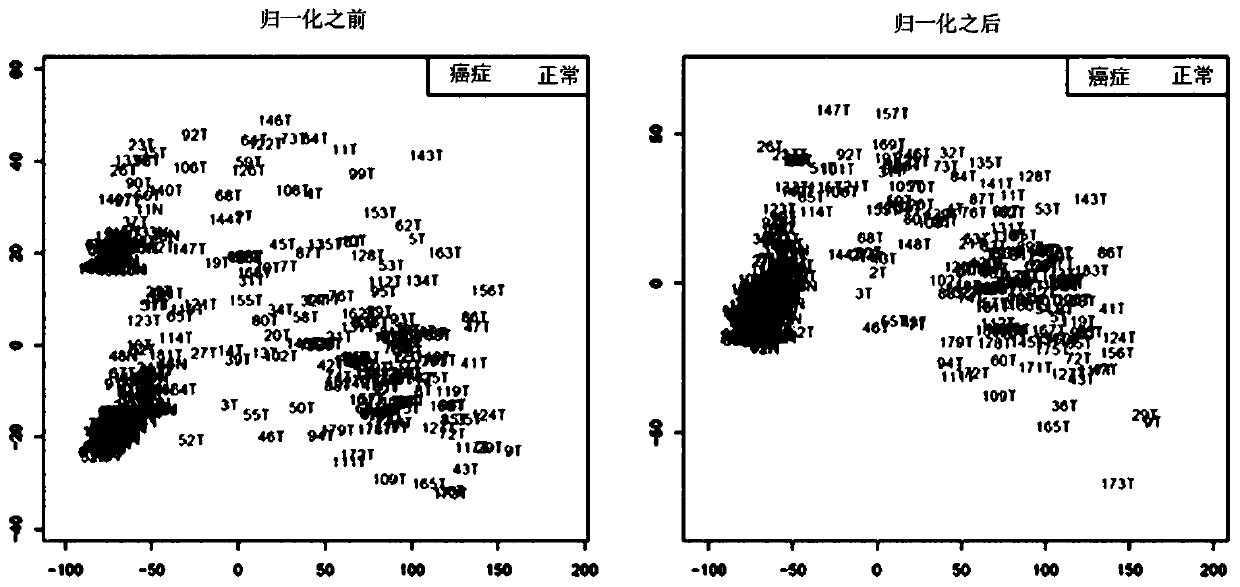
[0096] Methylation data was analyzed by a self-built pipeline. Probes with a low degree of methylation under normal conditions and a high degree of methylation in tumors were selected.
[0097] First, DMPs showing methylation differences between normal and cancer samples were selected.
[0098] Select 7 probes with very low methylation status in normal samples and very high methylation of more than 50% in more than 70% of cancer patients, and have machine learning methods to verify the efficiency ( figure 1 ,blue).
[0099] Select probes whose methylation is very low (below 10%) in normal samples and high on average above 30% in liver cancer patients, and perform machine learning to select the probes that can effectively distinguish liver cancer / norm...
Embodiment 3
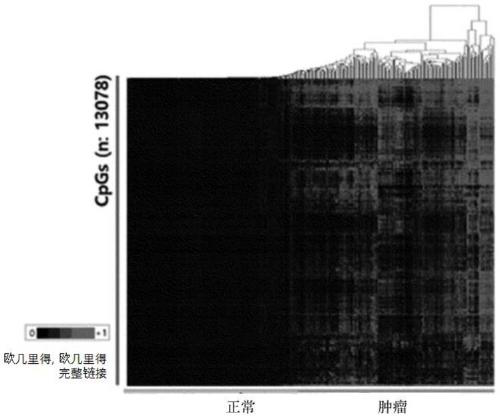
[0101] Example 3. Probe Selection by Heatmap
[0102] As a result of investigating the DNA methylation of 182 liver cancer samples and 127 normal samples, 100,053 DMP (differentially methylated probes, differently methylated probes) were selected from more than 5% of liver cancer samples that were overmethylated .
[0103] A very low 13,078 probes with methylation below 10% were selected from normal samples to enable blood biopsy in DMP showing differences between normal / cancer samples.
[0104] Among the selected probes, 7 probes with over 50% hypermethylation were selected from more than 70% of cancer patients (Table 1).
[0105] 【Table 1】
[0106] distinguish Probe ID Ratio of liver cancer with over 50% hypermethylation (%) Probe 1 cg20172627 78.16 Probe 2 cg22538054 77.59 Probe 3 cg27583690 74.14 Probe 4 cg19951303 72.99 Probe 5 cg22524657 71.84 Probe 6 cg24563094 70.11 Probe 7 cg25744484 70.11
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com