Expansion method for DNA methylation chip data
An extension method, a methylation technology, is applied in the direction of electrical digital data processing, special data processing applications, instruments, etc., and can solve problems that affect the practicability of algorithms, slow algorithm speed, and underutilization of methylation level data, etc., to achieve Excellent forecasting performance, improved forecasting speed, and accurate forecasting results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
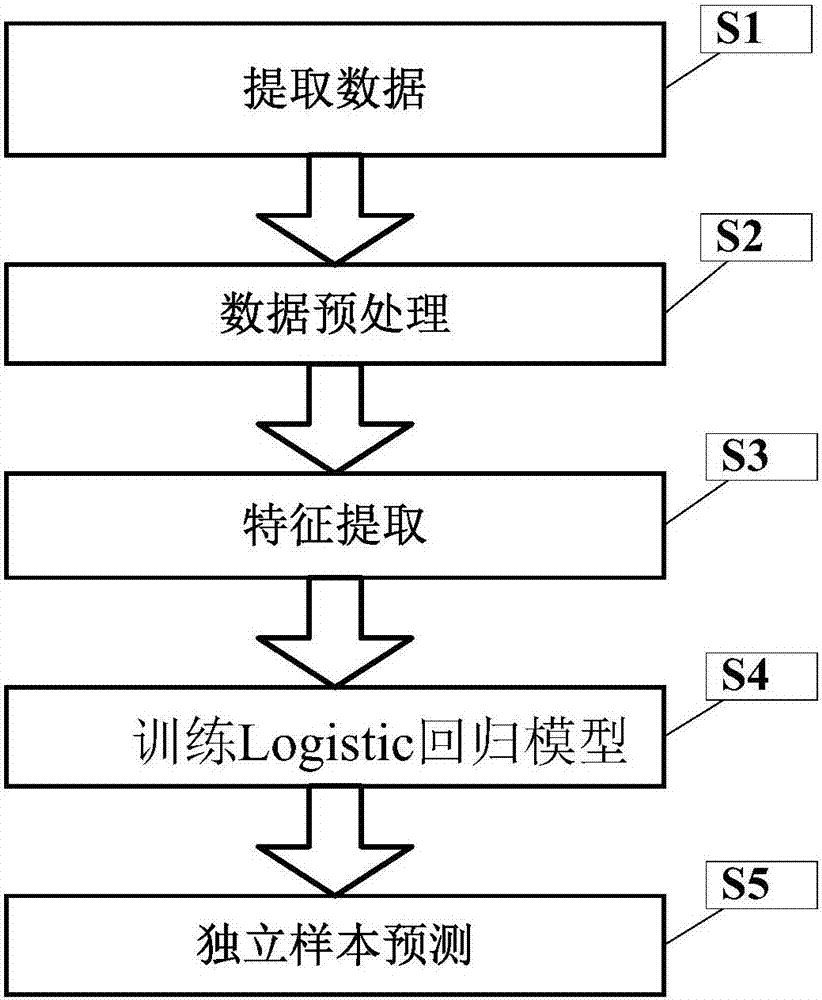
[0036] figure 1 It is a flow chart of a DNA methylation chip data expansion method of the present invention.
[0037] In this example, if figure 1 As shown, a method for extending DNA methylation chip data of the present invention comprises the following steps:
[0038] S1. Extract data
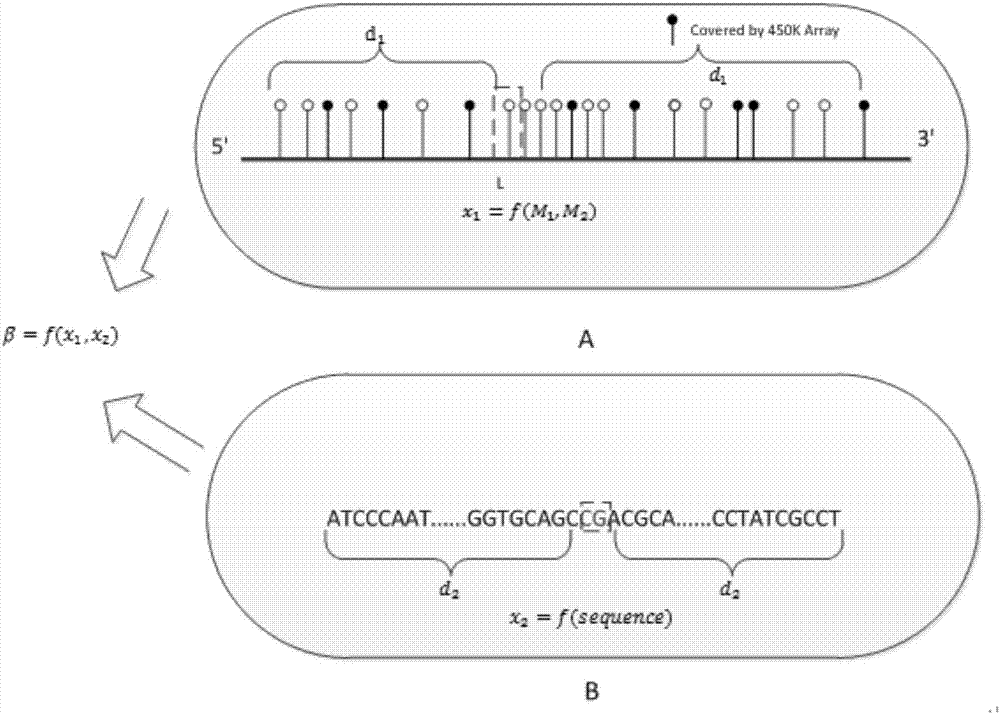
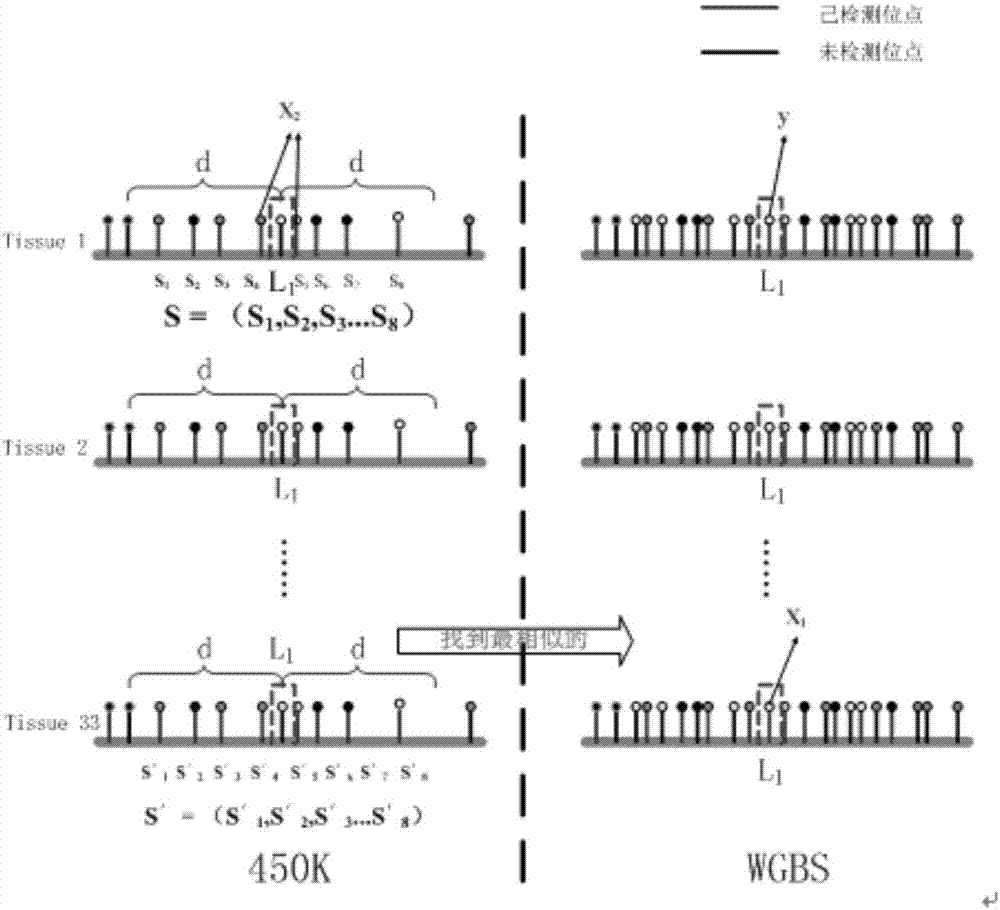
[0039] 31 different tissues T were obtained from the methylation public database GEO 1 , T 2 ,...,T 31 Whole-genome sulfite sequencing data and DNA methylation microarray data corresponding to 31 tissues; in addition, another arbitrary tissue T 32 Whole-genome sulfite sequencing data and DNA methylation microarray data.
[0040] S2, data preprocessing
[0041]Determine whether there is a null value in each row of the whole genome bisulfite sequencing data and the DNA methylation chip data, and if there is a null value, delete the corresponding row to obtain the standard whole genome bisulfite sequencing data and DNA methylation chip data. Basic chip data;
[0042] S3, feature extract...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com