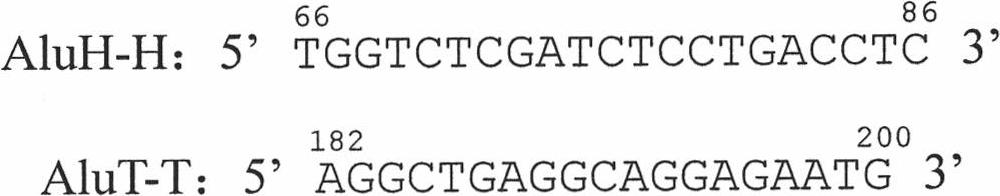Method for detecting characteristics of gene region based on inter-alu polymerase chain reaction
A chain reaction and polymerase technology, applied in the biological field, can solve problems such as the lack of in-depth research on the genome
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
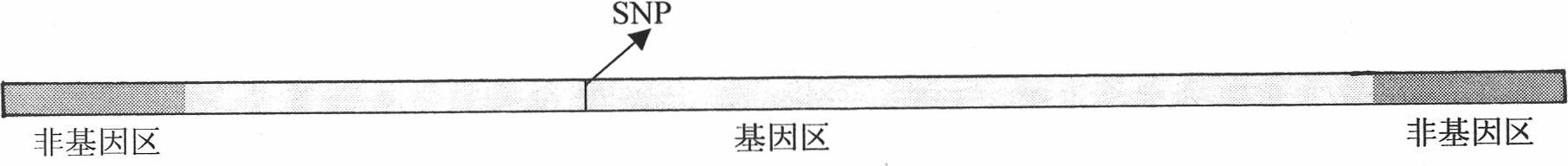
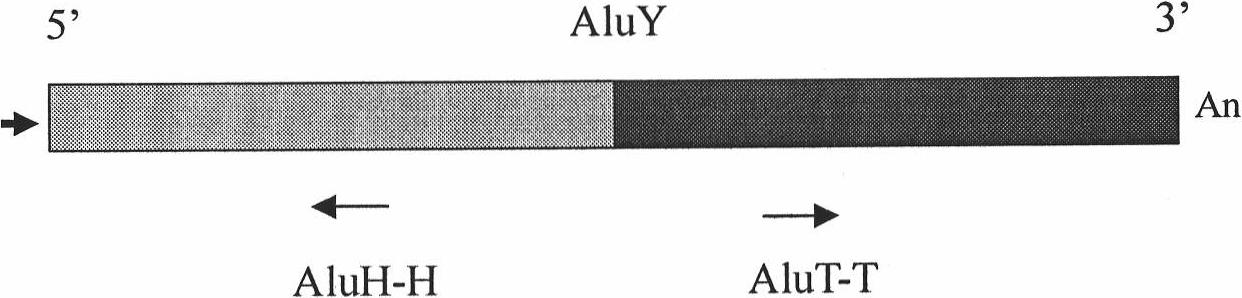
[0047] The detected SNP is the SNP concentrated in the gene region in the genome DNA of haploid or homozygous diploid or heterozygous diploid human. figure 1 An example of such a SNP within a gene region is shown. In order to obtain the sequence of the gene region, it is necessary to amplify the gene region intensively through inter-Alu PCR reaction, and then detect the SNP in the amplified gene region by next-generation sequencing technology. figure 2 Shown are the positions in the Alu structure of two oligonucleotide primers designed with small fragments in the AluY common sequence for inter-Alu PCR in the present invention, and the amplification directions of these two primers in genomic DNA; and image 3 Shown are the specific positions and sequences of these two primers. In inter-Alu PCR, through the action of thermostable DNA polymerase, a single oligonucleotide primer can be complementary to the detected template DNA at an appropriate sequence position to form a hybri...
Embodiment 2
[0051] Example 2 is similar to Example 1, the difference is that this example mainly focuses on the research on the correlation of polygenic diseases, using this method to detect the relationship between SNPs, insertions / deletions and diseases in gene regions, especially through the "exon "Capture" analyzes the relationship between SNPs, insertions / deletions and diseases in exons of gene regions. In order to obtain the phenotypic characteristics of tumor genome replication error (replication error) and more sequencing data, the sequence near the "3'" end of the AluY common sequence was selected as the oligonucleotide primer, and the primer amplification direction was "tail-to-tail" . Figure 5 Shown is the sequence of the oligonucleotide primer and its position in AluY. The positions of the oligonucleotide primers are from the 278th to the 295th in the Alu consensus sequence. The base is "5'-GAGCGAGACTCCGTCTCA-3'". Combine this oligonucleotide primer with the aforementioned o...
Embodiment 3
[0056] This embodiment also has similarity with embodiment 1 and embodiment 2. Because the vast majority of 5-methylcytosine (5mC) is mainly concentrated in the repeat sequence Alu family rich in CpG sites, and it is estimated that in the entire human genome, the Alu family can contain up to 33% of CpG sites . Previous studies have shown that in the tumor genome, the methylation level of some special CpG sites in the Alu sequence is significantly reduced, and this phenomenon is particularly prominent in the AluY subfamily. In addition, in the research on schizophrenia, we found that the methylation level of some CpG sites in and around the AluY sequence also changed significantly. Therefore, in this embodiment, in order to study the change degree of the methylation level of the CpG site in the gene region in various diseases, the positions of the selected AluY oligonucleotide primers need to be adjusted. Because the "C" in unmethylated CpG sites will be converted to "T" afte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com