Long non-coding RNA (lncRNA) marker derived from adipocyte exosomes and application and product of lncRNA marker
A technology of adipocytes and exosomes, applied in the field of biomedicine, can solve problems such as exosomes without TUG1 gene, and achieve the effect of improving prognosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Induce adipocyte formation in vitro, and use PA to stimulate lipid droplet hypertrophy, and simulate the process of adipocyte hypertrophy in vitro.
[0074] 1. Induction and differentiation of 3T3-L1 preadipocyte cell line
[0075] 1) Inoculate 3T3-L1 preadipocytes on a culture plate, and use high-glucose DMEM medium containing 10% fetal bovine serum at 37°C, 5% CO 2 to cultivate.
[0076] 2) After the cells reach 80-90% confluence and confluence for 2 days (fusion means to let the cells contact and inhibit, that is, change the medium after confluence to allow them to grow for another two days), add the final concentration of 0.5mmol / L IBMX (stock solution concentration 0.5mmol / L), dexamethasone at a final concentration of 1umol / L (stock solution concentration of 1mg / ml (2.5mmol / L)) and insulin at a final concentration of 10ug / ml in 10% fetal bovine serum Incubate in high-sugar DMEM for 48 hours; (the inducer is then added to the culture medium and mixed evenly before...
Embodiment 2
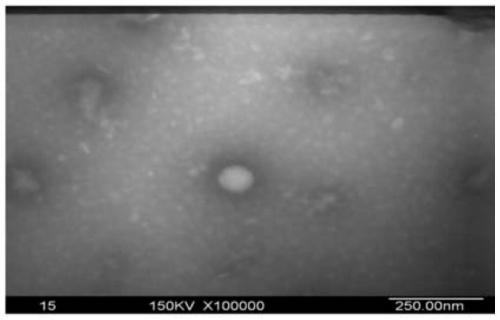
[0096] After exosomes were extracted from the supernatant of adipocytes, they were identified under the electron microscope.
[0097] 1. Supernatant Collection
[0098] 1) 3T3-L1 preadipocytes were induced with PA for 24 hours after adipogenesis, and BSA was used as a control.
[0099] 2) After 24 hours, replace the culture medium with serum-free DMEM medium.
[0100] 3) After 24 hours, the supernatant was collected and stored in a -80 refrigerator for later use.
[0101] 2. Exosome Extraction
[0102] 1) The supernatant after induction with equal volumes of PA and BSA was taken out from the -80 refrigerator and thawed on ice.
[0103] 2) After the supernatant is filtered through a 0.22um pore size filter, it is concentrated using a 100K ultrafiltration tube.
[0104] 3) Add ExoQuick-TC Separation Reagent to the concentrated supernatant at a ratio of 1:5, mix by inverting, and let stand overnight in a refrigerator at 4°C.
[0105] 4) Take out the concentrated supernatant ...
Embodiment 3
[0121] After extracting exosomes from adipocyte supernatant, nanoparticle tracking analysis and identification.
[0122] Exosome extraction method is the same as in Example 2
[0123] Nanoparticle Tracking Analysis (NTA) is to track and analyze the Brownian motion of each particle, and calculate the hydrodynamic diameter and concentration of nanoparticles in combination with the Stockes-Einstein equation. NTA technology has been recognized by the exosome research field as one of the exosome characterization methods; this part is assisted by the School of Chemistry, Shandong University.
[0124] Experimental results:
[0125] As attached in the manual Figure 5 and Figure 6 As shown, in the supernatant induced by PA and BSA, after exosomes were collected, nanoparticle size analysis showed that the peak particle size distribution was within 200 nm, which was consistent with the physiological characteristics of exosomes.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap