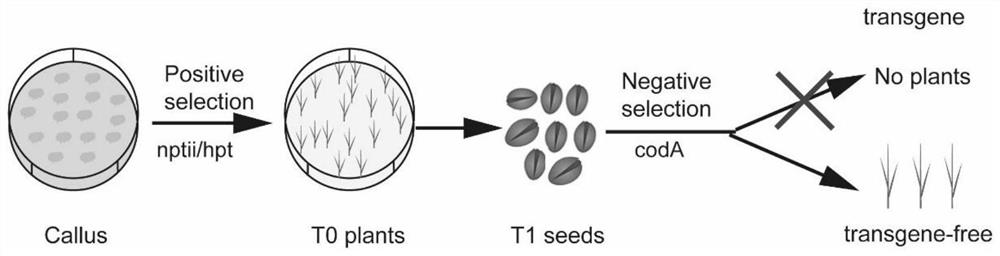
A method for rapid screening of non-transgenic site-directed mutation plants
A non-transgenic, site-directed mutation technology, applied in botany equipment and methods, biochemical equipment and methods, plant products, etc., can solve the problems of increasing manpower input, reduce time and labor, and avoid false positive effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
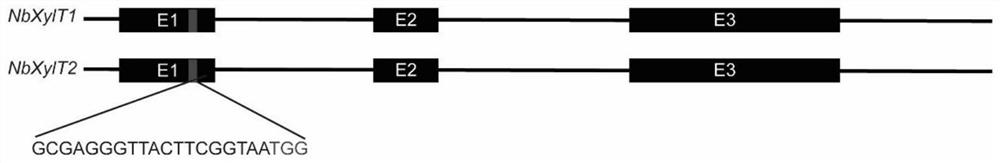
[0031] In this embodiment, a mutant of the non-transgenic tobacco Nbxylt gene was screened using positive and negative screening markers, and the process was as follows:
[0032] 1. Construction of positive and negative selection markers (coda::nptii or hpt::coda) suitable for tobacco knockout vectors
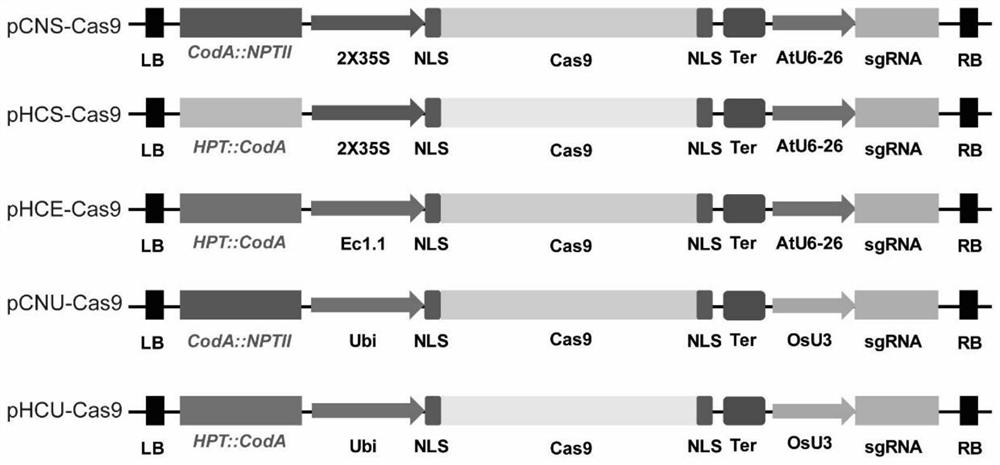
[0033](1) Construction of pCNS-Cas9 vector: it contains coda::nptii positive and negative selection markers, 35S promoter drives the expression of Cas9, and AtU6-26 promoter drives the expression of sgRNA, which is suitable for tobacco gene knockout. Design primers 411-F SEQ ID NO.3: 5'-AGTAGATGCCGACCGGATCTGTC-3' and RB-R SEQ ID NO.4: 5'-TGACAGGATATATTGGCGGGTAAAC-3', using pCambia1300 as a template (pCambia1300 can be purchased from Youbao Biological Company), and expand Fragment 1 containing the backbone of the Agrobacterium vector was amplified; primers RB-F SEQ ID NO.5: 5'-GTTTACCCCGCCAATATATCCTGTCA-3' and 411-R SEQ ID NO.6: 5'-ATCTCATTGCCCCCCGGATC-3' were designed, using pH...
Embodiment 2
[0059] In this example, the Arabidopsis thaliana AtBRI1 gene was site-directedly mutated using a positive and negative selection marker vector containing hpt::coda, and the process was as follows:
[0060] 1. Construction of the positive and negative selection marker hpt::coda knockout vector pHCE-Cas9 suitable for Arabidopsis
[0061] pHCE-Cas9 vector: contains hpt::coda positive and negative selection markers, EC1.1 promoter to drive the expression of Cas9, and AtU6-26 promoter to drive the expression of sgRNA, suitable for gene knockout in Arabidopsis. Design primers 411-F SEQ ID NO.30: 5'-AGTAGATGCCGACCGGATCTGTC-3' and RB-R SEQ ID NO.31: 5'-TGACAGGATATATTGGCGGGTAAAC-3', using pCambia1300 as a template to amplify the Agrobacterium vector backbone Fragment 1; design primers RB-F SEQ ID NO.32: 5'-GTTTACCCCGCCAATATATCCTGTCA-3' and hyg-R SEQ ID NO.33: 5'-CTTTGCCCTCGGACGAGTGCTGG-3', using pHEE401 as a template (pHEE401 plasmid: in the literature "Hui -LiXing, Li Dong, Zhi-Ping ...
Embodiment 3
[0081] In this example, a vector containing a positive and negative selection marker (coda::nptii or hpt::coda) is used to site-directed mutation of the rice Osglossy2 gene, and the process is as follows:
[0082] 1. The construction of a knockout vector containing positive and negative selection markers (coda::nptii or hpt::coda) suitable for rice
[0083](1) Construction of the pCNU-Cas9 vector: it contains coda::nptii positive and negative selection markers, the maize Ubi-1 promoter drives the expression of Cas9, and the OsU3 promoter drives the expression of sgRNA, which is suitable for gene knockout in rice. Design primers 411-F SEQ ID NO.41: 5'-AGTAGATGCCGACCGGATCTGTC-3' and RB-R SEQ ID NO.42: 5'-TGACAGGATATATTGGCGGGTAAAC-3', using pCambia1300 as a template to amplify a fragment containing the backbone of the Agrobacterium vector 1; Design primers RB-F SEQ ID NO.43: 5'-GTTTACCCCGCCAATATATCCTGTCA-3' and 411-R SEQ ID NO.44: 5'-ATCTCATTGCCCCCCGGATC-3', using pHUN411 as a te...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap