Tags and methods for simultaneous visualization of gene dna, mrna and protein in living cells
A living cell and labeling technology, which is applied in the field of DNA, mRNA and protein labeling, can solve the problems of limited application, lack, and inability to clearly capture the quantitative analysis of nascent RNA, and achieve the effect of expanding the scope of application and accurate quantification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] S1. Design and construct a blue fluorescent protein sequence containing introns:
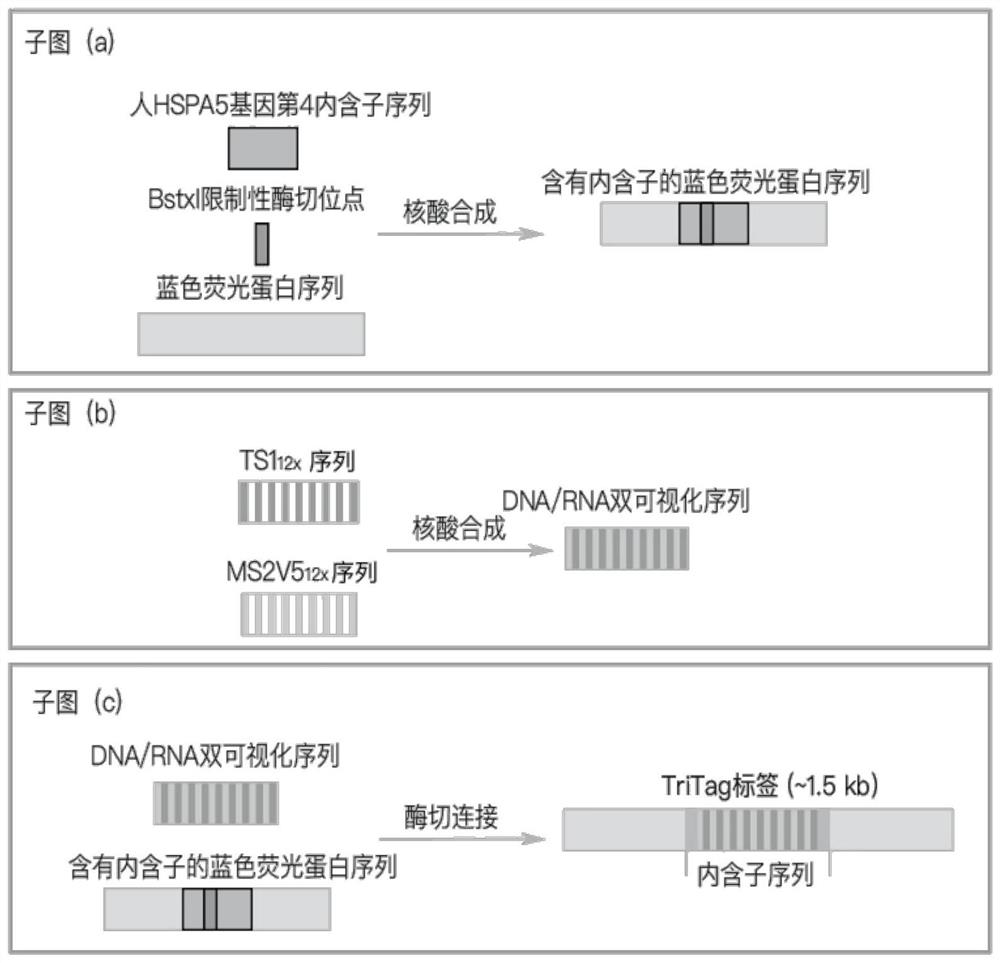
[0068] The human HeLa cell genome was extracted, and the blue fluorescent protein sequence containing introns was obtained by nucleic acid synthesis, such as figure 1 as shown in (a);
[0069] S2. The screened TS1 capable of efficiently targeting and editing the nematode genome 12x Sequence with reduced repetitiveness of MS2V5 12x The sequence is synthesized by nucleic acid, and the TS1 repeat unit and the MS2V5 repeat unit are alternately connected to form a DNA / RNA dual visualization sequence, such as figure 1 as shown in (b);
[0070] S3. Construction of TriTag tags: Insert the dual visualization sequence into the intron region of the blue fluorescent protein sequence containing introns to obtain TriTag tags using the enzyme-cut ligation method, such as figure 1 As shown in (c), specifically the BstxI restriction enzyme site inserted into the intron sequence.
Embodiment 2
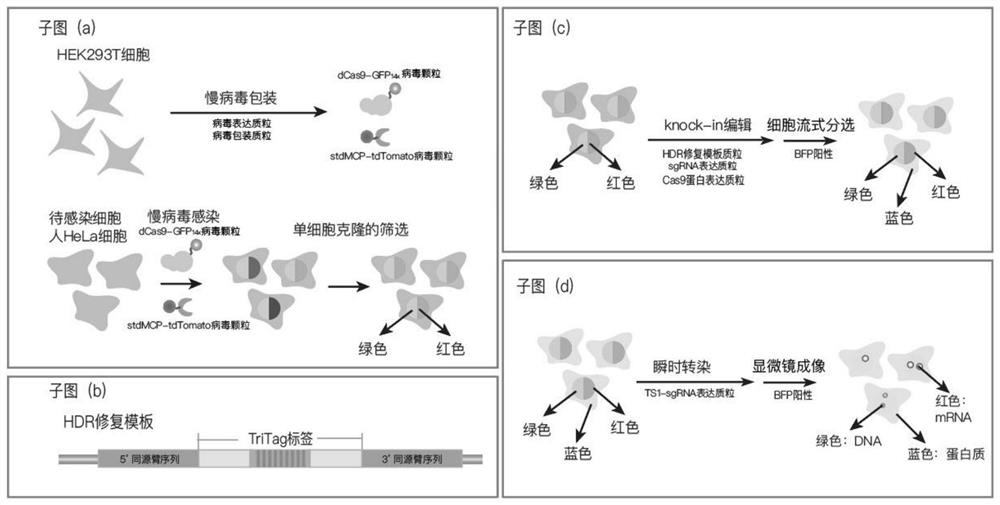
[0072] (1) Construct a visualized stable cell line, such as figure 2 As shown in (a):
[0073] (1.1) On the same day, HEK293T cells were cultured with PS-free medium and spread into a 12-well plate to form the first cell culture medium. The concentration of HEK293T cells added could make the cell density reach more than 80% the next day;
[0074] (1.2) The next day, carry out lentiviral packaging: use 75 μl of serum-reduced medium (Opti-medium) to premix each 750ng of dCas9-GFP 14xAfter virus expression plasmid and stdMCP-tdTomato virus expression plasmid, pCMV-dR8.91 plasmid of 705ng and PMD2.G plasmid of 87ng, then add 4.5μl Fugene transient transfection reagent (Promega) to form liposome, add dropwise to the In the supernatant of the first cell culture medium, HEK293T cells in the first cell culture medium were transiently transfected to prepare cells containing virus particles;
[0075] (1.3) After 12 hours of transient transfection in step (1.2), suck off the supernata...
Embodiment 3
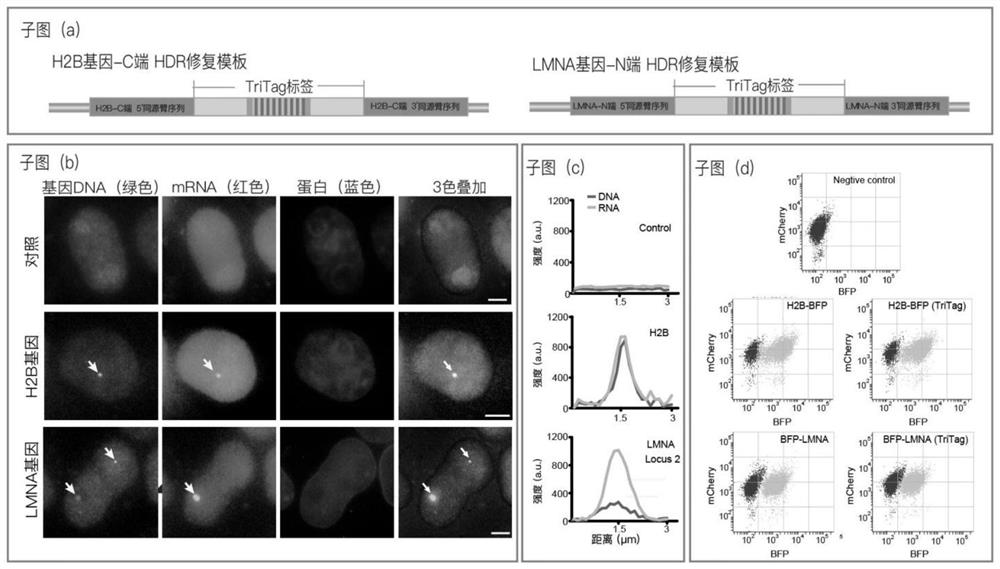
[0100] (1) Construct the target endogenous gene (human histone H2B gene and nuclear membrane protein LMNA gene) visualization system in a stable cell line and transfect and sort:
[0101] (1.1) According to the protein structure prediction of the target endogenous gene product H2B histone and LMNA nuclear membrane protein, choose to insert the TriTag tag into the C-terminus of the human histone H2B gene, and insert the TriTag tag into the N of the human nuclear membrane protein LMNA gene end. The human HeLa cell genome was extracted, and the homology arm sequences (5' homology arm sequence and 3' homology arm sequence) on both sides of the H2B gene C-terminus or LMNA gene N-terminus were respectively amplified, and the length of the homology arm sequence was about 500 bp. The HDR repair template of 5' homology arm sequence-TriTag tag-3' homology arm sequence was constructed by homologous recombination. Such as image 3 (a) shown.
[0102] (1.2) As in steps (2.2)-(2.4) in Ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com