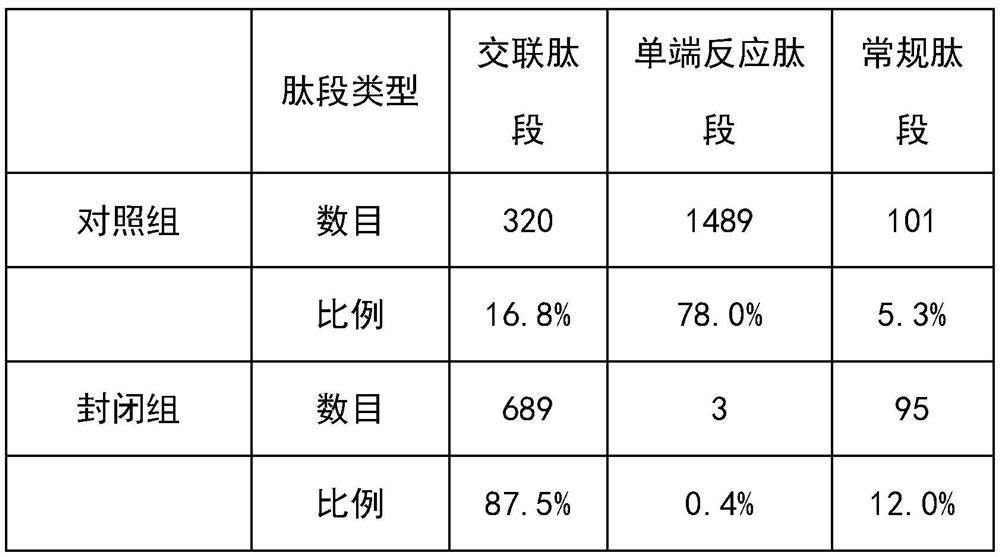
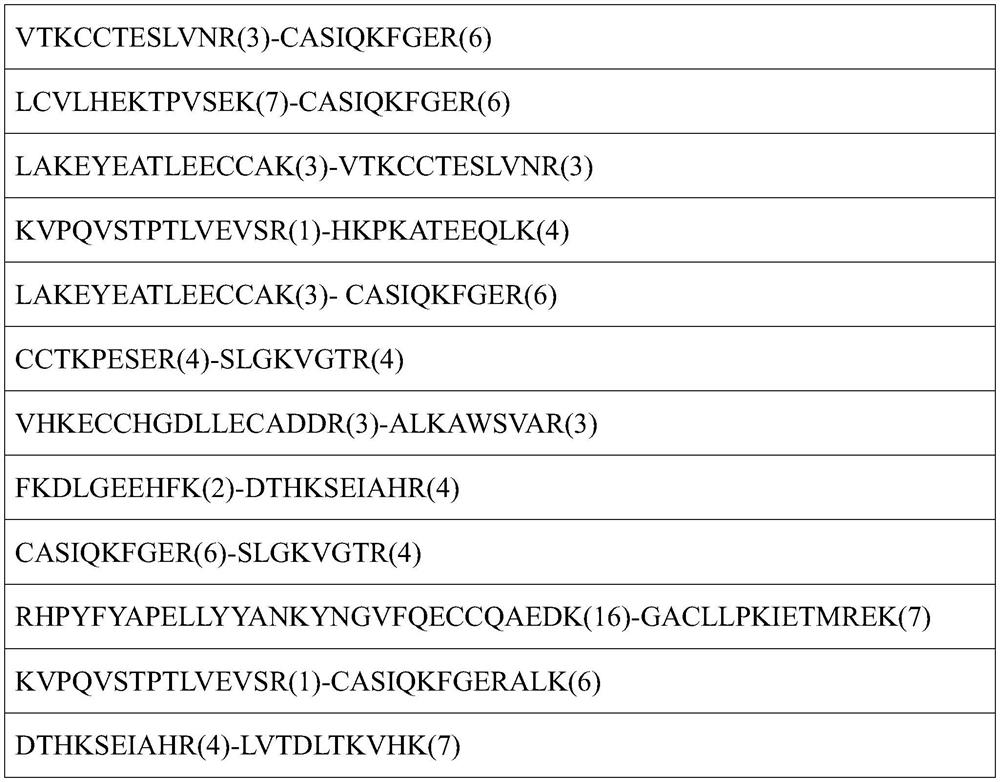
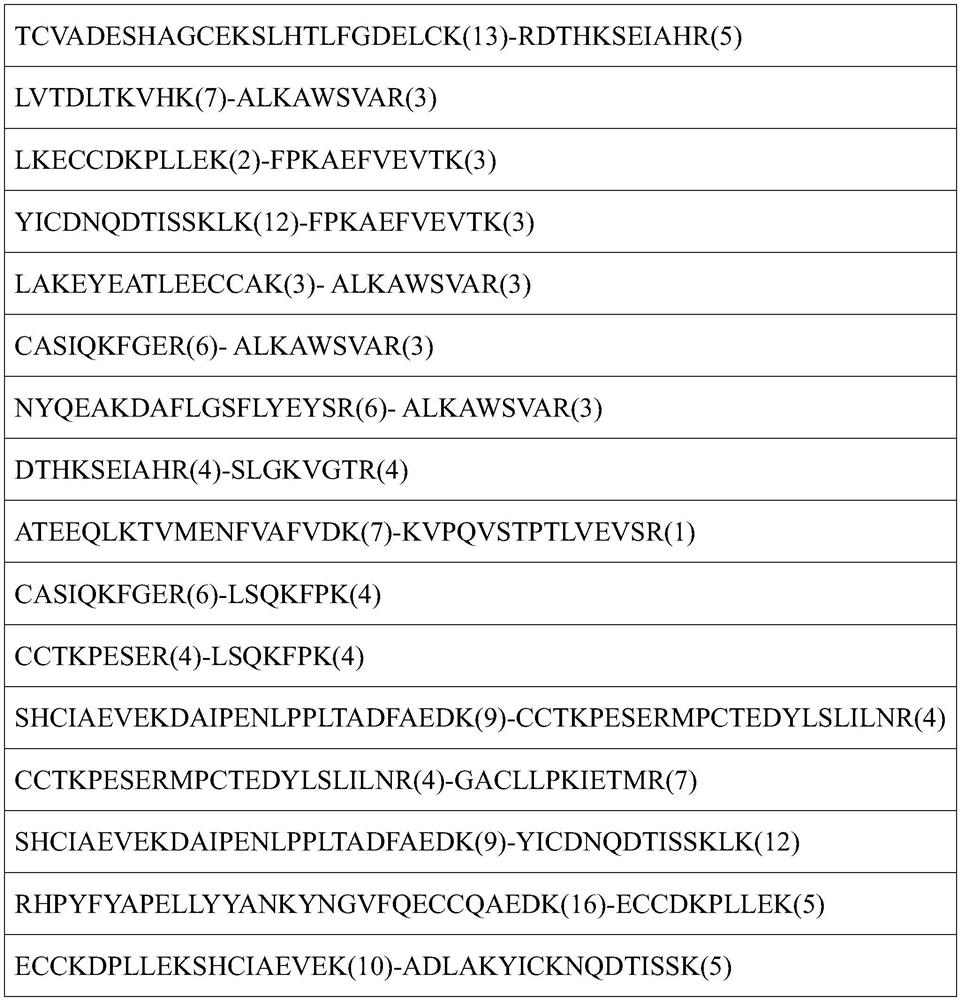
A single-end cross-linked peptide removal method and its application to the analysis of cross-link sites in protein complexes
A protein and protein technology, applied in the analysis of materials, material analysis by electromagnetic means, biological testing, etc., can solve the problem of low abundance of cross-linked peptides, complex cross-linked samples, and the inability to achieve selective enrichment of cross-linked peptides and other problems, to achieve the effect of combining fast and efficient, high enrichment selectivity, and improved identification sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] 1. Cross-linking reaction of protein samples
[0026] Dissolve 10 μg of bovine serum albumin sample (BSA) in 20 mM 4-hydroxyethylpiperazineethanesulfonic acid buffered saline solution (HEPES) at pH 7.4 to a final protein concentration of 1 mg / mL and use dimethyl sulfoxide (DMSO) Propargyl nitroheptyl disuccinimidyl ester (BSPNO) was prepared at a concentration of 25 mM, and the cross-linking agent was added to the BSA solution to make the final concentration 1 mM, and reacted at room temperature for 1 h.
[0027] 2. Blocking of unreacted NHS groups
[0028] Add amino-PEG at a final concentration of 10 mM 3 -Biotin, react at room temperature for 2h.
[0029] 3. Click chemistry
[0030] Add 20mM CuSO to the sample 4 , 160mM THPTA and 500mM Vc to make the final concentrations of 100μM, 800μM and 2.5mM respectively, add 20mM Diazo Biotin-Azide to make the final concentration of 200μM, and react at 60°C for 2h.
[0031] 4. Protein precipitation
[0032] Add four times ...
Embodiment 2
[0051] 1. Cross-linking reaction of protein samples
[0052] Dissolve 10 μg of bovine serum albumin sample (BSA) in 20 mM 4-hydroxyethylpiperazineethanesulfonic acid buffered saline solution (HEPES) at pH 7.4 to a final protein concentration of 1 mg / mL and use dimethyl sulfoxide (DMSO) Azidonitroheptyl disuccinimide ester was prepared at a concentration of 25 mM, the cross-linking agent was added to the BSA solution so that the final concentration was 1 mM, and the reaction was carried out at room temperature for 1 h.
[0053] 2. Blocking of unreacted NHS groups
[0054] Add amino-PEG at a final concentration of 10 mM 3 -Biotin, react at room temperature for 2h.
[0055] 3. Click chemistry
[0056] Add 20mM CuSO to the sample 4 , 160mM THPTA and 500mM Vc to make the final concentrations of 100μM, 800μM and 2.5mM respectively, add 20mM Alkyne Biotin-Azide with a final concentration of 200μM, and react at 60°C for 2h.
[0057] 4. Protein precipitation
[0058] Add eight ti...
Embodiment 3
[0079] 1. Cross-linking reaction of protein samples
[0080] Dissolve 10 μg of rabbit creatine kinase protein sample (CK) in 50 mM phosphate-buffered saline (PBS) with a pH of 7.4, the final protein concentration is 1 mg / mL, and use dimethylformamide (DMF) to prepare 25 mM propargyl nitric acid Heptyl disuccinimidyl ester (BSPNO), the cross-linking agent was added to the CK solution so that the final concentration was 1 mM, and reacted at room temperature for 1 h.
[0081] 2. Blocking of unreacted NHS groups
[0082] Add amino-PEG11-biotin at a final concentration of 10 mM, and react at room temperature for 2 h.
[0083] 3. Click chemistry
[0084] Add 20mM CuSO4, 160mM BTTAA and 500mM Vc to the sample to make the final concentrations of 100μM, 800μM and 2.5mM respectively, add 20mM Diazo Biotin-Azide with a final concentration of 200μM, and react at 37°C for 1h.
[0085] 4. Protein precipitation
[0086] Add four times the volume of pre-cooled acetone to the sample, and p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com