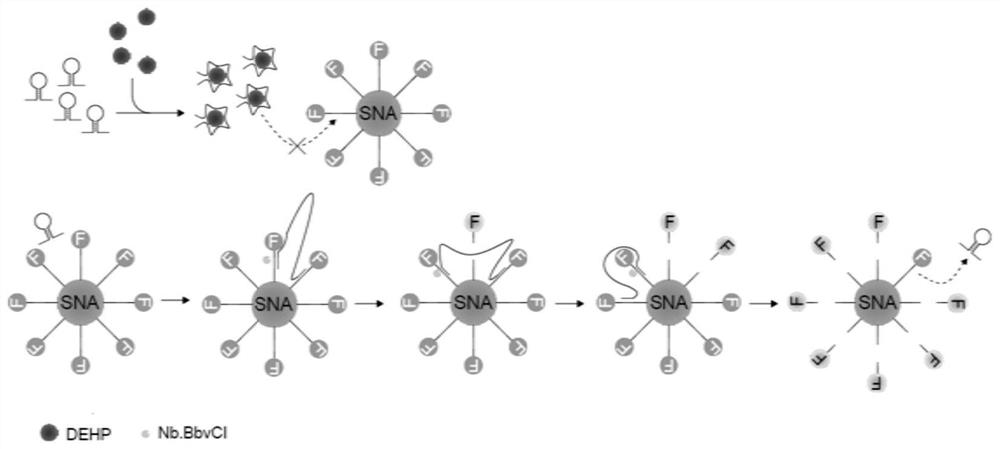
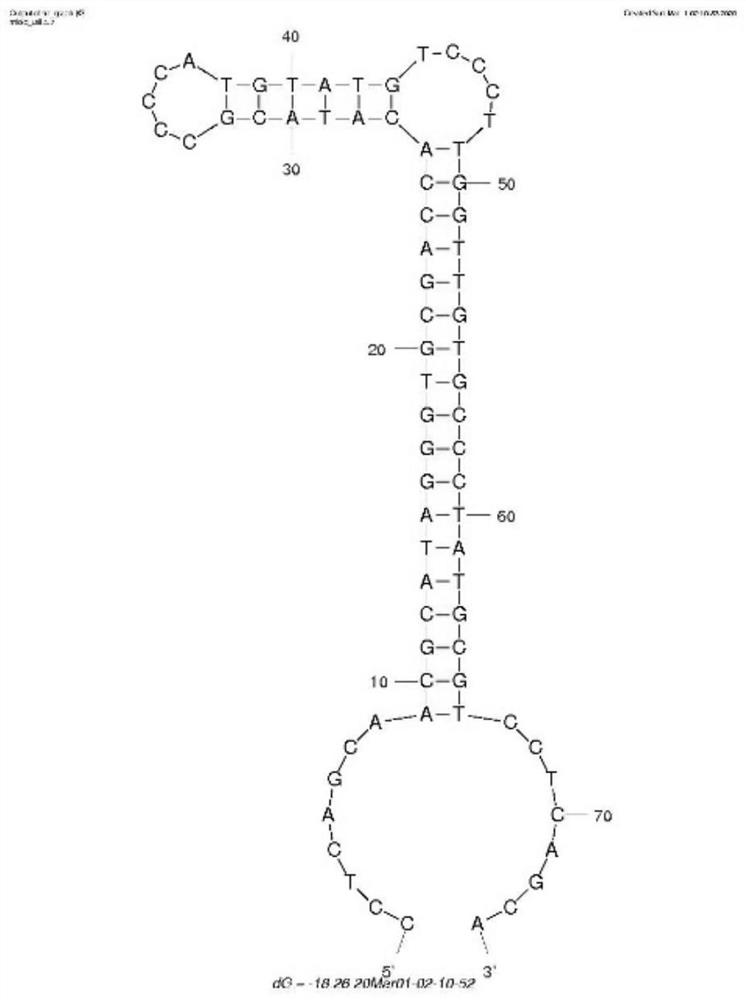
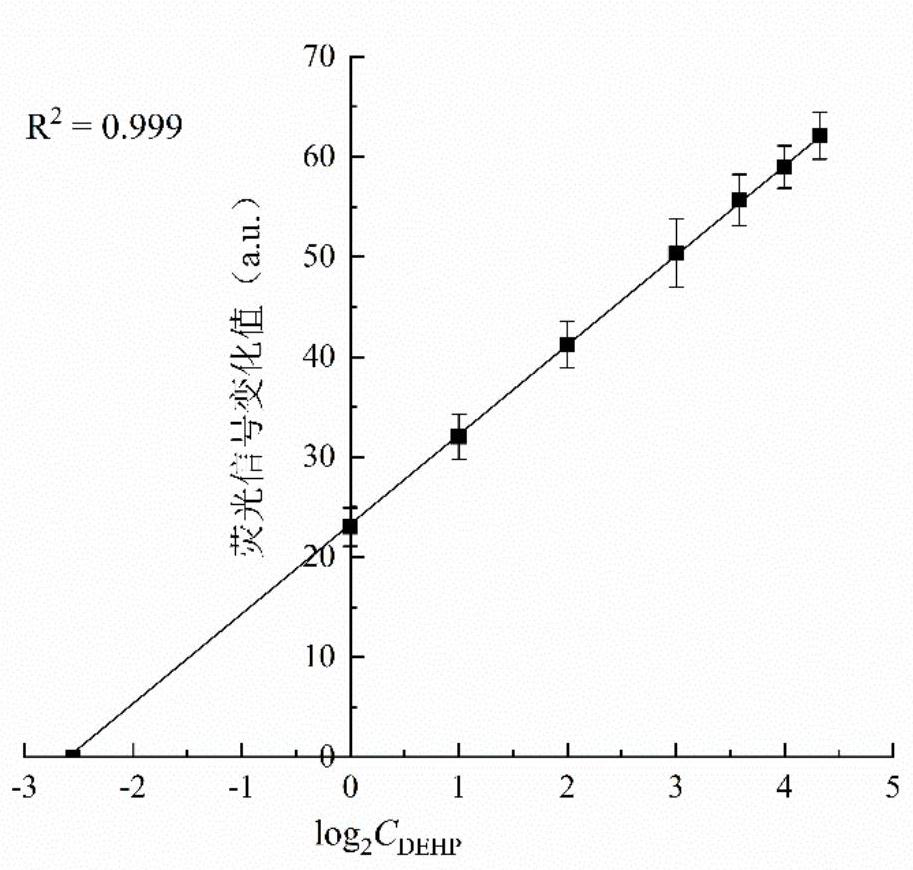
Fluorescence detection method for bis (2-ethyl) hexyl phthalate based on coupling of nucleic acid aptamer with spherical nucleic acid
A technology of phthalic acid di- and nucleic acid aptamers, which is applied in fluorescence/phosphorescence, biological testing, measuring devices, etc., to achieve the effects of fast and portable on-site detection, high controllability, and short preparation time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] A fluorescent detection method based on nucleic acid aptamer coupling spherical nucleic acid di(2-ethyl)hexyl phthalate, comprising the following steps:
[0048] (1) Preparation of gold nanoparticles
[0049] In a 500 mL three-necked flask, heat 100 mL of 1 wt% HAuCl under vigorous stirring 4 The solution was boiled, and then 5mL, 38.8mM sodium citrate solution was added thereto, stirred vigorously, and reacted for 15min. After the solution changed from pure yellow to wine red, the heat source was removed and continued to cool to room temperature to obtain a gold nanoparticle solution. Store in the dark at 4°C for later use;
[0050] (2) Preparation of spherical nucleic acid
[0051] Dissolve the functional oligonucleotide chain in dithiothreitol phosphate buffer (pH=8), let it stand at room temperature for 2 hours, and then use NAP-5 column to elute to remove impurities; 50 μM functional oligonucleotide after removal Mix the nucleotide chain with step (1) 100 μL gol...
Embodiment 2
[0058] A fluorescence detection method based on nucleic acid aptamer coupling spherical nucleic acid di(2-ethyl)hexyl phthalate, the difference from Example 1 is that the amount of each substance in the test sample of the two The content varies and specifically includes the following steps:
[0059] (1) Preparation of gold nanoparticles
[0060] In a 500 mL three-necked flask, heat 100 mL of 1 wt% HAuCl under vigorous stirring 4 The solution was boiled, then 5mL, 38.8mM sodium citrate solution was added thereto, stirred vigorously, and reacted for 15min. After the solution changed from pure yellow to wine red, the heat source was removed, and continued to cool to room temperature to obtain a gold nanoparticle solution in 4 Store in dark at ℃ for later use;
[0061] (2) Preparation of spherical nucleic acid
[0062] Dissolve the functional oligonucleotide chain in dithiothreitol phosphate buffer (pH=8), let it stand at room temperature for 2 hours, and then use NAP-5 column ...
Embodiment 3
[0069] A fluorescence detection method based on nucleic acid aptamer coupling spherical nucleic acid di(2-ethyl)hexyl phthalate, the difference from Example 1 is that the amount of each substance in the test sample of the two The content is slightly different and includes the following steps:
[0070] (1) Preparation of gold nanoparticles
[0071] In a 500 mL three-necked flask, heat 100 mL of 1 wt% HAuCl under vigorous stirring 4 The solution was boiled, and then 5mL, 38.8mM sodium citrate solution was added thereto, stirred vigorously, and reacted for 15min. After the solution changed from pure yellow to wine red, the heat source was removed and continued to cool to room temperature to obtain a gold nanoparticle solution. Store in the dark at 4°C for later use;
[0072] (2) Preparation of spherical nucleic acid
[0073] Dissolve the functional oligonucleotide chain in dithiothreitol phosphate buffer (pH=8), let it stand at room temperature for 2 hours, and then use NAP-5 ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap