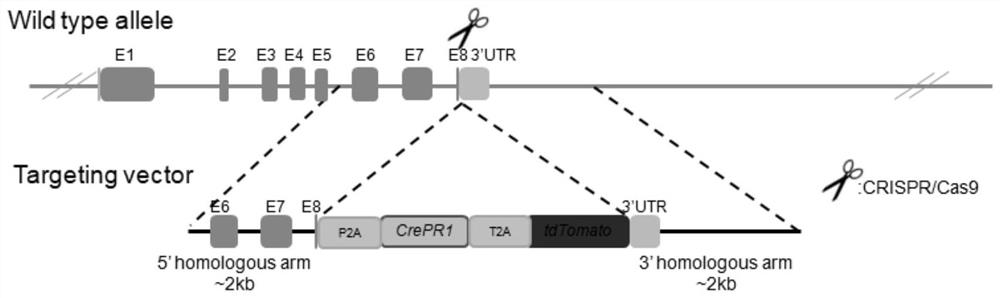
Construction and application of KRT10 site-directed gene knock-in P2A-CrePR1-T2A-tdTomato mouse model
A KRT10, gene knock-in technology, applied in the field of bioengineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1, verifying the DNA sequence of the constructed strain mouse.
[0051] Select C57BL / 6J mice as donor mice, using the primer pair: EGE-STY-061-MSD-F:5'-GCATCAAGCTTGGTACCGATGCTGAAACTA-3'(SEQ ID NO.11) and EGE-STY-061-MSD-R : 5'-ACTTAATCGTGGAGGATGATCTCATACTGGT-3' (SEQ ID NO.12), carry out PCR amplification and perform PCR amplification and sequencing verification on the mouse tail target site sequence of C57BL / 6J, to ensure that the DNA sequence of the constructed strain mouse can be compared with The subsequent design of the sgRNA recognition sequence is compatible.
[0052] After sequencing, it was confirmed that the length of the amplified product of this example was 763bp, which was completely consistent with the DNA sequence of C57BL / 6J mice.
Embodiment 2
[0053] Embodiment 2, design and acquisition of sgRNA.
[0054] According to the target site sequence known in Example 1, 7 potential sgRNAs were designed, which are:
[0055] sgRNA1: 5'-AGTGATCAGGACGATTATTGAGG-3' (SEQ ID NO.11);
[0056] sgRNA2: 5'-AGTGATCAGGACGATTATTGAGG-3' (SEQ ID NO.12);
[0057] sgRNA3: 5'-AGTCTCTTCATACGGGCCACTGG-3' (SEQ ID NO.1);
[0058] sgRNA4: 5'-GACGAAAGGACTCTACCCTCAGG-3' (SEQ ID NO.13);
[0059] sgRNA5: 5'-AATAATCGTCCTGATCACTTTGG-3' (SEQ ID NO.14);
[0060] sgRNA6: 5'-CATAGAAGAGTCTCTTCATACGG-3' (SEQ ID NO.15);
[0061] sgRNA7: 5'-TACTAACAAGCTATTACAAA AGG-3' (SEQ ID NO. 16).
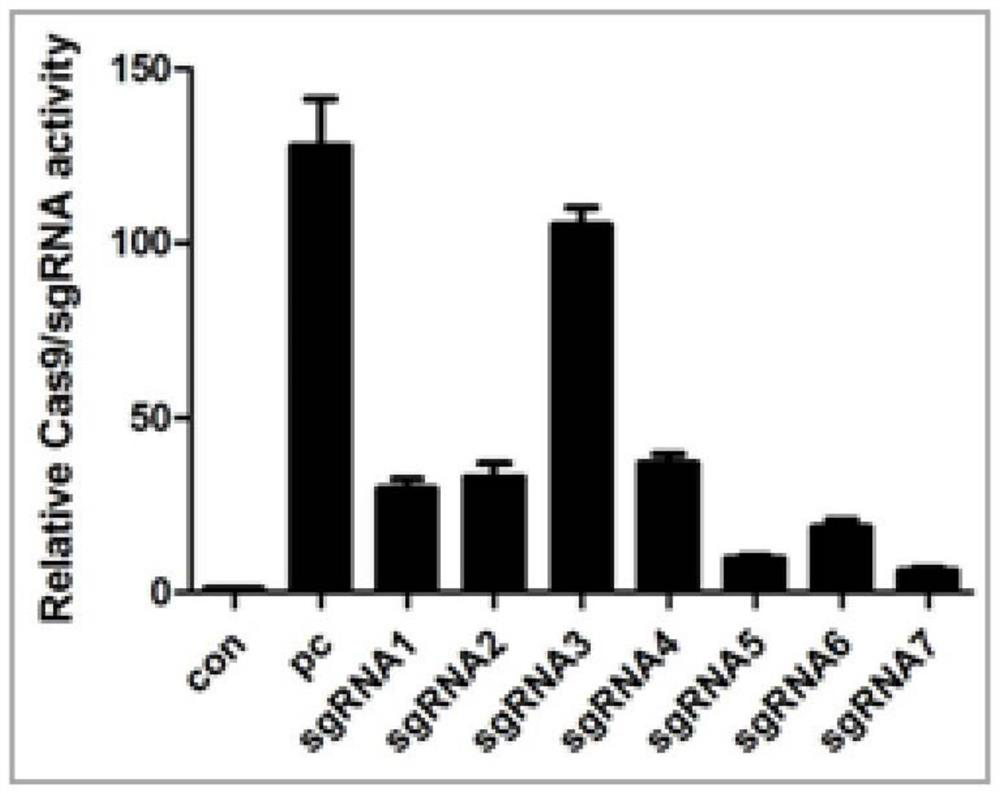
[0062] Biocytogen’s self-developed CRISPR / Cas9 activity detection method-UCATM method was used to detect the activity of sgRNA, and the detection results were as follows: figure 2 As shown, the activity of sgRNA3 is the highest, and the sequence of sgRNA3 is used as sgRNA for subsequent construction.
[0063] The sgRNA obtained by screening was connected to a plasmid vec...
Embodiment 3
[0064] Embodiment 3, construction Cas9 / sgRNA plasmid.
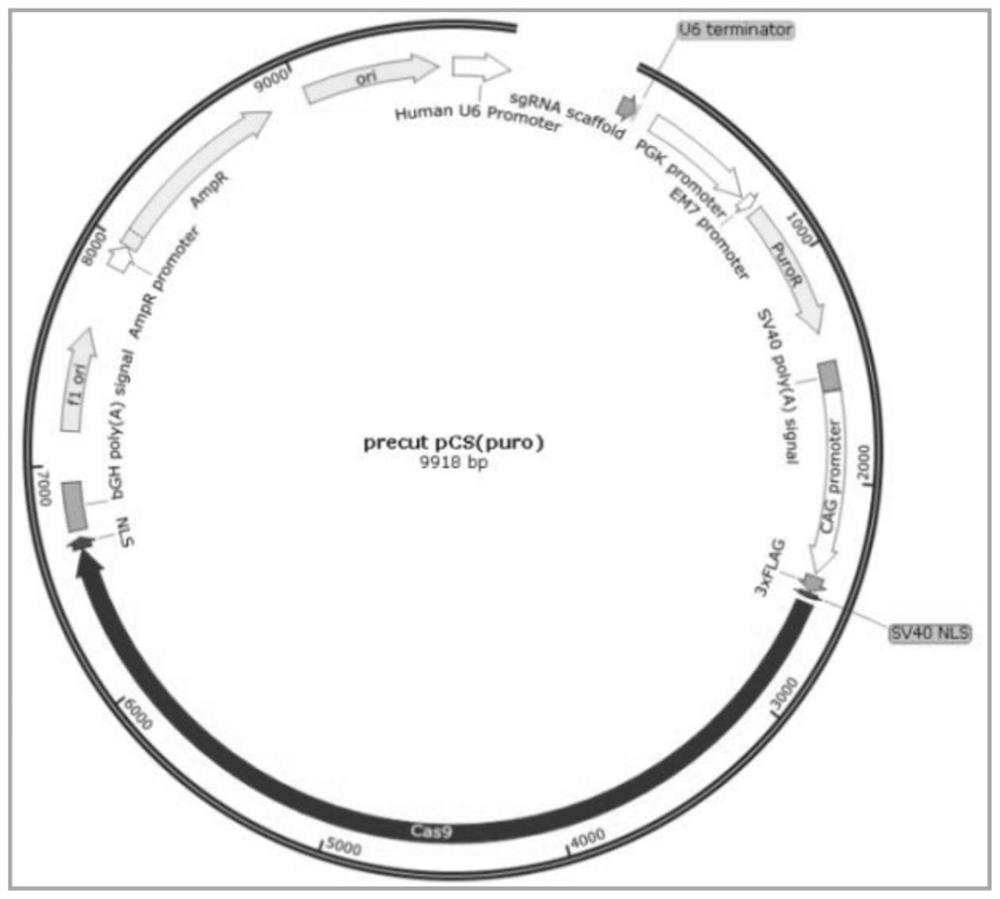
[0065] Synthesize oligos (oligonucleotides) through the sgRNA sequence designed in Example 2, and then connect the sgRNA into the pCS vector by annealing and polymerization, and the ligation product will be sent for correct sequencing after conversion; wherein the size of the Cas9 / sgRNA plasmid vector is 9918bp, the map is as follows image 3 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com