A dna recombination site prediction method based on xgboost
A DNA recombination and prediction method technology, applied in the field of computational biology, can solve the problems of sequence limitation, low efficiency, time-consuming, etc., and achieve the effect of increasing recombination rate, improving prediction efficiency, and high visibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] In order to clearly illustrate the technical solutions of the present invention, the following specific embodiments are combined with the appendix Figure 1-4 To illustrate the present invention, the examples herein are only used to explain the present invention, but not to limit the present invention.
[0029] It should be noted that the following detailed description is exemplary and intended to provide further explanation of the invention. Unless otherwise defined, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs.
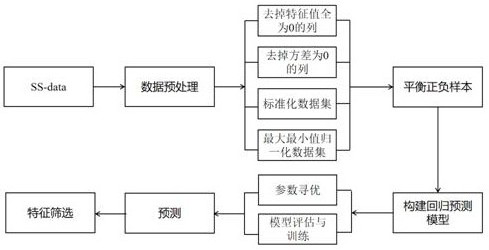
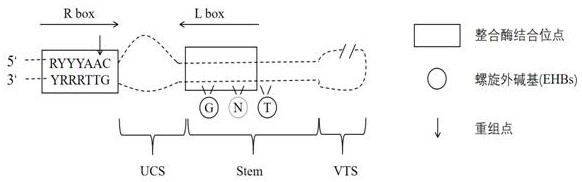
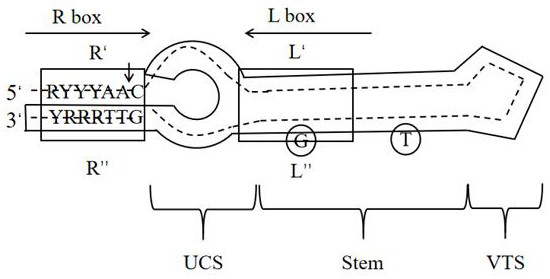
[0030] figure 1 The workflow steps of XGBattCPred's method for predicting DNA recombination sites are shown. The DNA recombination site selected in this example is the attC site of the bacterial integration system, and the structural diagram of the attC site is as follows figure 2As shown, since the structure of this site is highly dependent ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com