Process
A motor protein and polynucleotide technology, applied in biochemical equipment and methods, instruments, analysis materials, etc., can solve the problem that polynucleotides cannot be accurately determined
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
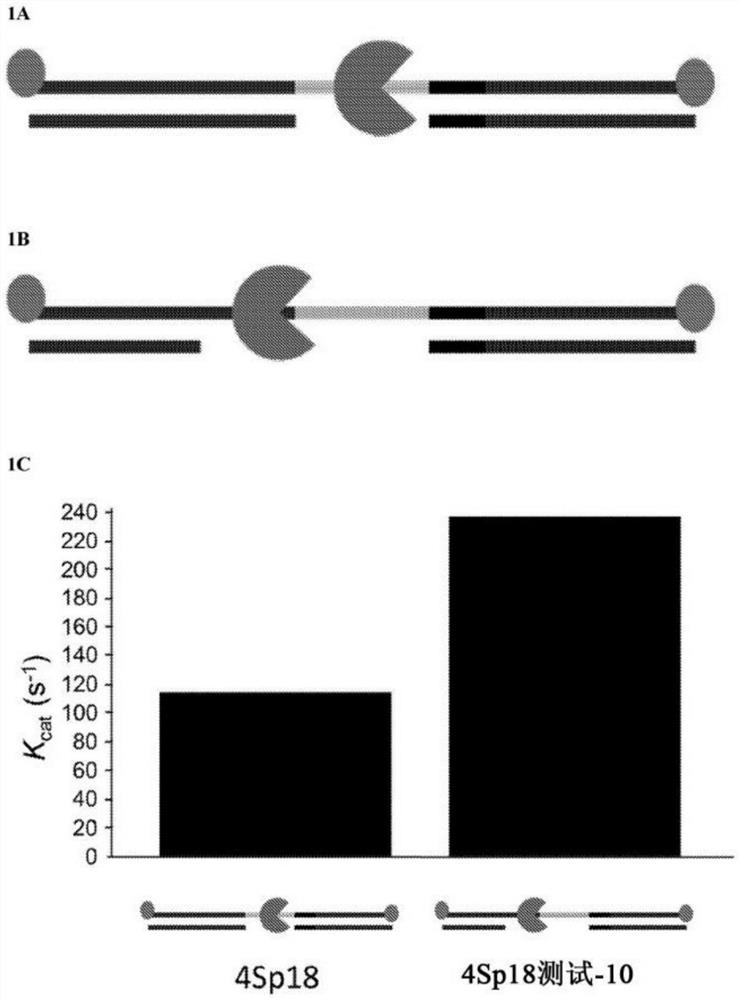
[0397] This example demonstrates that useless turnover is reduced when the motor protein is stalled on the spacer according to the methods disclosed herein compared to when the motor protein is stalled near the spacer.
[0398] A first DNA construct comprising a single-stranded polynucleotide strand of SEQ ID NO:9 and comprising a spacer (4iSp18 unit, IDT) was combined with complementary single-stranded DNA strands (SEQ ID NO:10 and SEQ ID NO:11) hybridize. SEQ ID NO: 10 is complementary to the portion of SEQ ID NO: 9 between the 5' end of SEQ ID NO: 9 and the 5' end of the spacer and is referred to as the "test" strand. The motor protein (blocks T4 Dda helicase) stalls on the spacer. The construct is in figure 1 Schematically depicted in A.
[0399] The second DNA construct was generated with a blocked T4 Dda helicase loaded on the spacer. The second DNA construct was identical to the first DNA construct except that SEQ ID NO:12 was used instead of SEQ ID NO:10. SEQ ID N...
Embodiment 2
[0403] This example compares adapters with various spacer designs and demonstrates that ineffective ATP turnover is reduced when motor proteins are stalled on the spacer according to the methods provided herein.
[0404] As shown in the results table below, a number of different adapters were made using the spacer design. The adapters tested comprised the following polynucleotide strands: (1) top strand, (2) bottom strand, and (3) closed strand. The top strand contains the spacer design being tested flanked by polynucleotide sequences on both sides. A closed strand is a closed portion of a single stranded polynucleotide as described herein.
[0405] adapter sequence
[0406] Top chain:
[0407] 33333333333333333333333333333CCTTTTTTTTGGCGTCTGCTTGGGTGTTTAACC / [spacer sequence] / CCACAACTTCGTTCAGTTACGTATTGCT (ie, SEQ ID NO: 13; SEQ ID NO: 14 and SEQ ID NO: 16 comprising the spanning spacer sequence. SEQ ID NO: 14 comprising SEQ ID NO: 15)
[0408] Bottom strand: 5phos / GCAATACG...
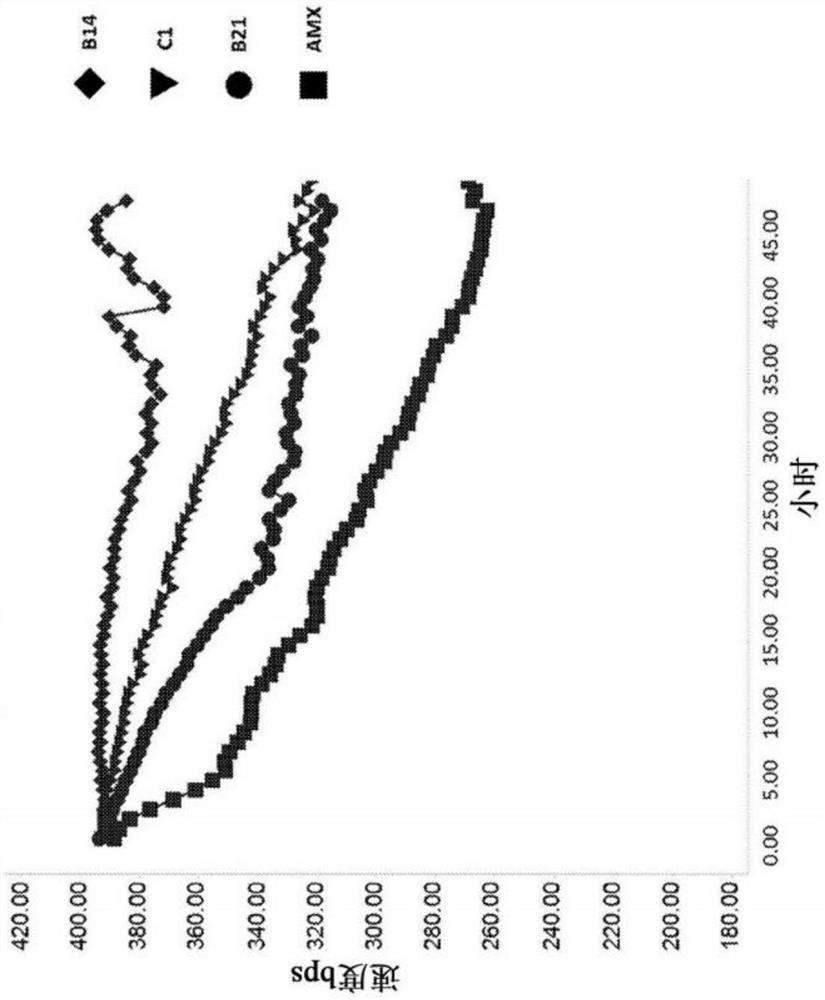
Embodiment 3
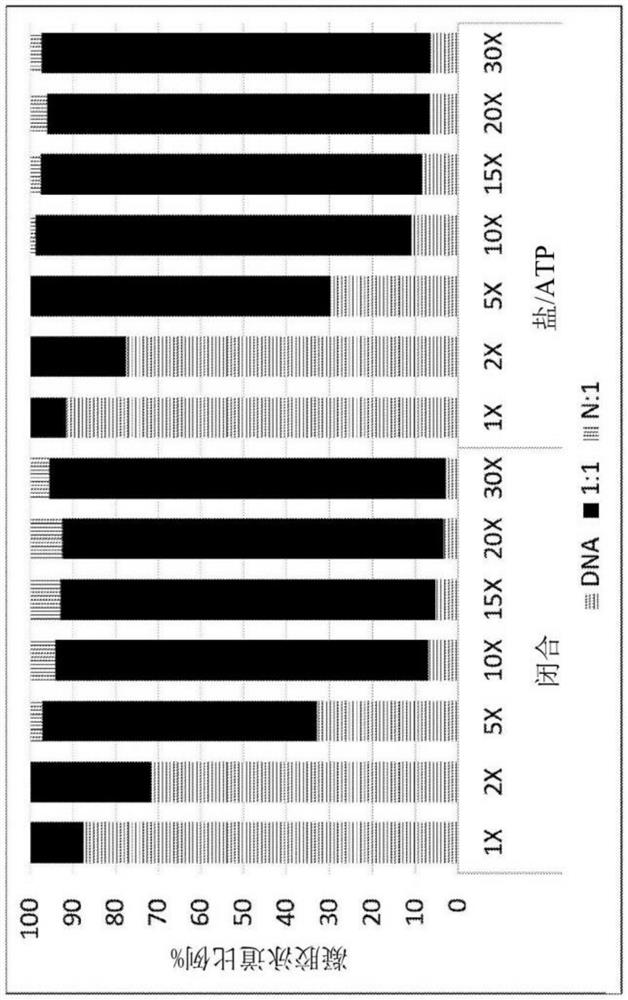
[0455] This example demonstrates that motor proteins can be loaded directly onto the spacer region of a polynucleotide adapter when the blocking portion ("closed strand") is already in place.
[0456] Polynucleotide adapters were prepared with top strand, bottom strand and closed strand sequences as described above in Example 2. The top strand comprises a spacer sequence comprising a plurality of nucleotide islands, such as described herein.
[0457] Adapter-containing polynucleotide strands were assembled by annealing the strands in potassium acetate buffer (400 mM HEPES pH 8.0, 400 mM potassium acetate) at the following concentrations:
[0458] Volume (μL) final concentration top chain 20 10μM closed chain 22 11μM bottom chain 22 11μM 1x KOAc buffer 100 0.5x nuclease free water 36 - total 200
[0459] The motor protein (Dda helicase) was mixed with annealed adapters at a final concentration of 50-500 nM DNA and...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Extinction coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com