Methods for assessing and treating cancer
An oncogene, farnesyltransferase technology, used in the evaluation and treatment of cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0116] Example 1: Microarray Analysis
[0117] From Zarnestra diluted with PBS and centrifuged with Ficoll-3,5-bis(acetylamino)-2,4,6-triiodobenzoic acid (1.077g / ml) TM FTI (active ingredient, (R)-6-[amino(4-chlorophenyl)(1-methyl- 1H -Imidazol-5-yl)methyl]-4-(3-chlorophenyl)1-methyl- 2(1H) -quinolinone)-treated patients, bone marrow samples were collected before and after treatment. Leukocytes were washed twice with PBS, resuspended in FBS containing 10% DMSO, and immediately frozen at -80°C. The samples were thawed at 37°C and 10 x volume of RPMI with 20% FBS was added dropwise over a period of 5 minutes. Cells were centrifuged at 500 g for 10 minutes and resuspended in 10 ml PBS containing 2 mM EDTA and 0.5% BSA. Samples were then passed through a 70 [mu]M filter (Becton Dickinson Labware, Franklin lakes, NJ) to remove any cell clumps. Cell viability was determined by trypan blue dye exclusion assay. The mean viability of bone marrow samples at thaw was 35% (range ...
Embodiment 2
[0125] Example 2: Leukemia cell antigen expression
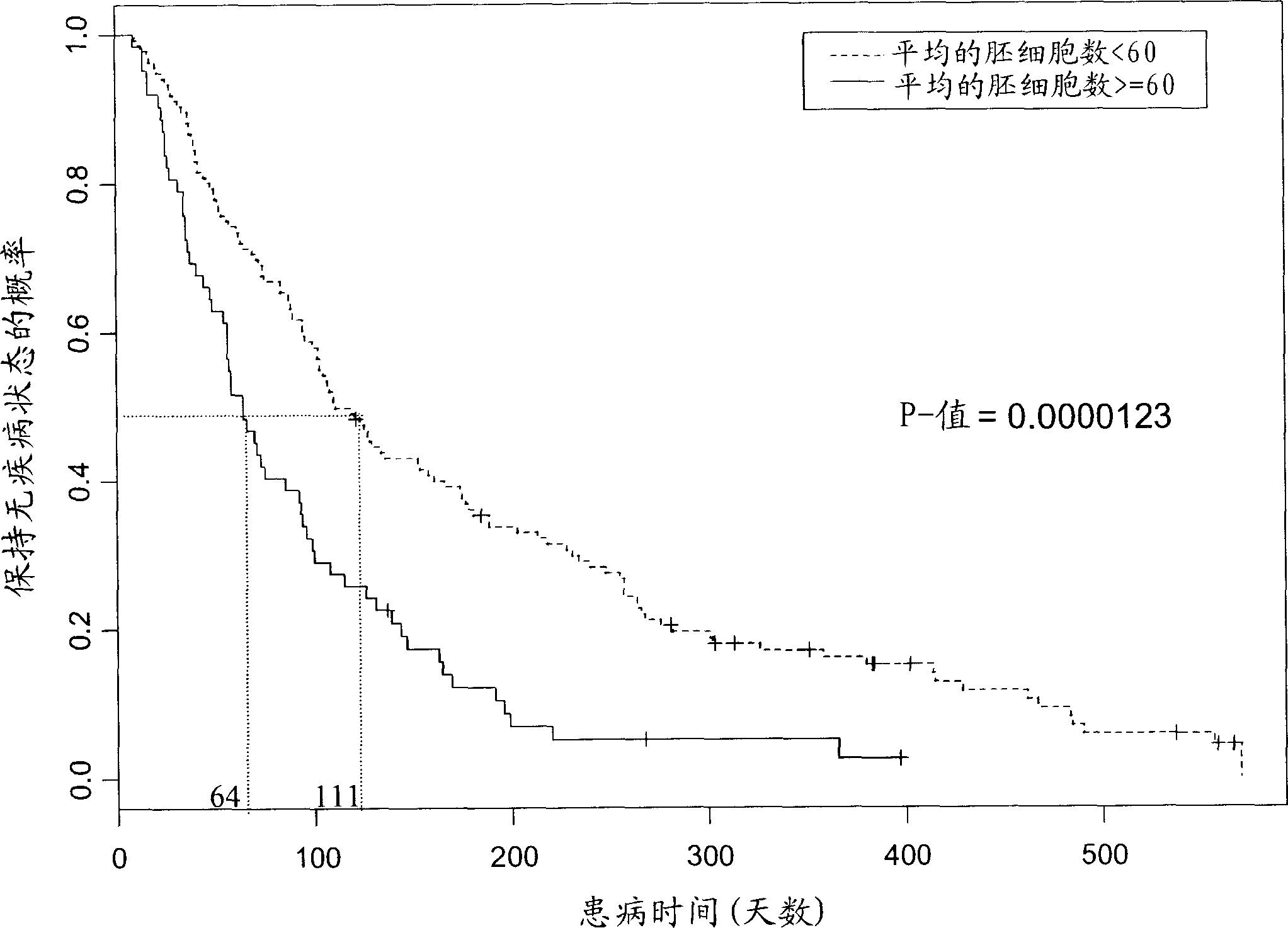
[0126] Leukemic blast cells are known to express the surface antigens CD33 and CD34. 95% and 55% of patients' myeloid leukemia cells were positive for CD33 and CD34, respectively. The average percentage of antigen-expressing cells in each sample was 13% for CD34 and 43% for CD33. Chi-squared analysis was performed to investigate the correlation between antigen expression levels and patient outcomes. Cutoffs of 15% and 60% were chosen for CD34 and CD33 levels, respectively. Low expression of CD33 and CD34 (less than about 15% of the CD34 cell population positive and less than about 60% of the CD33 cell population positive) was associated with patient response to Zarnestra TM Correlation (p=0.137, p=0.052, respectively). High expression of both antigens (greater than about 15% positive for the CD34 cell population and less than about 60% positive for the CD33 cell population) was also positively correlated with high blast ...
Embodiment 3
[0127] Example 3: Leukemia blast count analysis
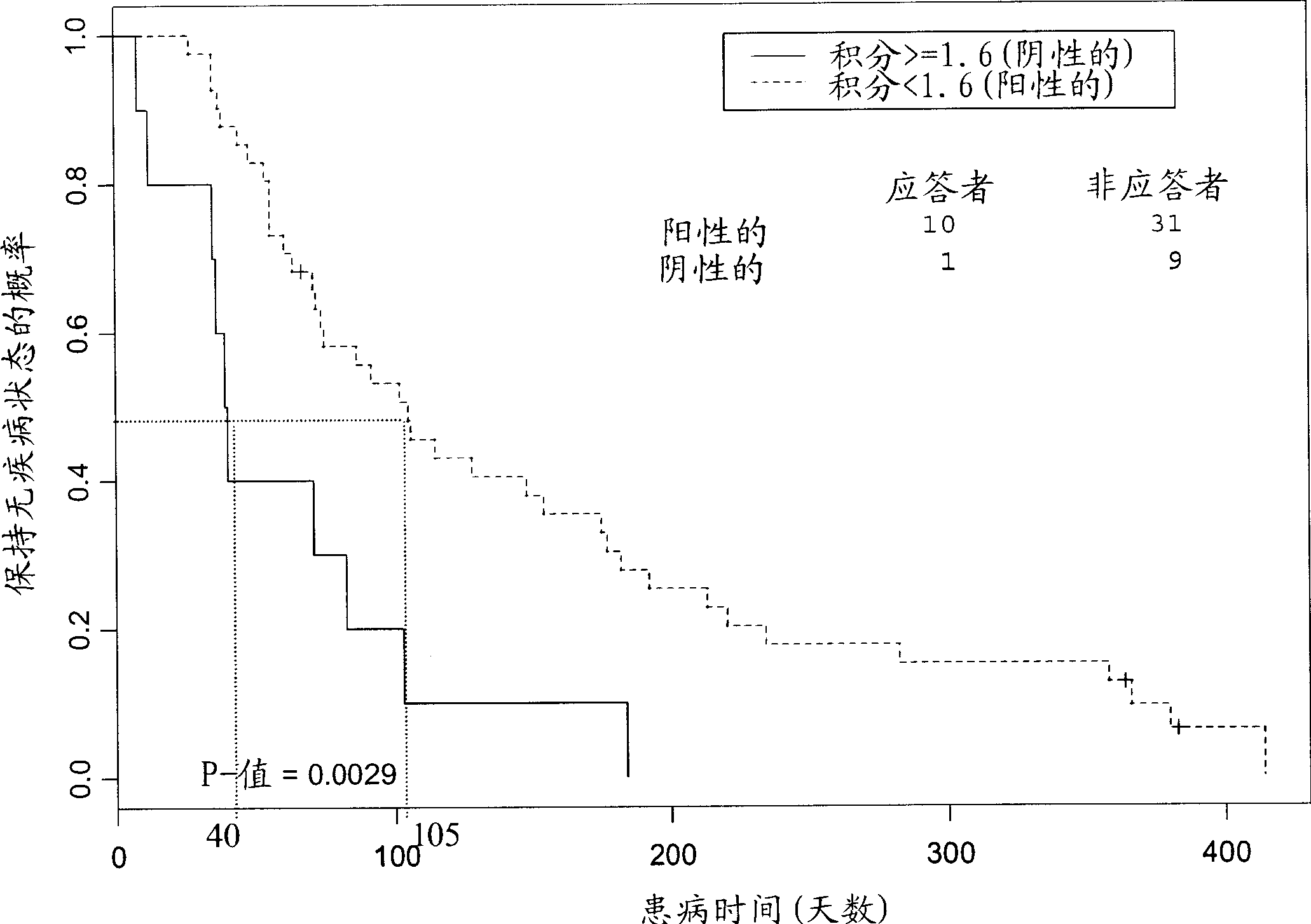
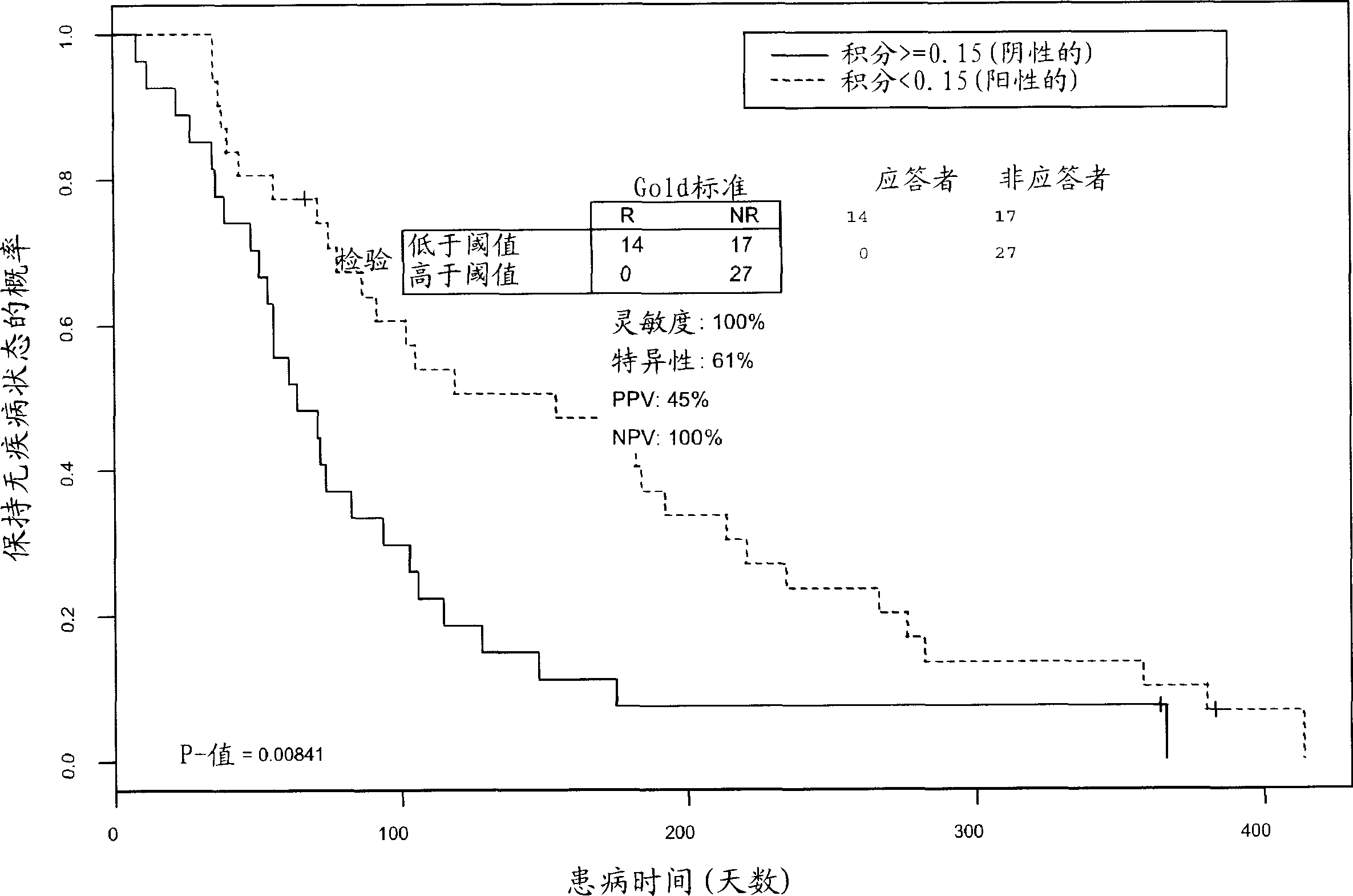
[0128] Analysis of CD33 and CD34 antigen expression suggested that leukemic blasts were associated with patient response. Means of sites and DCL blast count measurements were calculated and a Student's t-test was performed to investigate whether blast counts were associated with patient response in AML patients. Of a total of 199 patients evaluated, 24 had CR, CRp, PR, or SD, and the percentage of blasts correlated significantly with patient output (p=0.0006). Responders had lower numbers of blast cells (average 34%) than patients with progressive disease (average 51%). Only one of a total of 24 responders (defined as having SD) had a blast count above 60%. Chi-square analysis of all evaluable patients also found a significant correlation between high blast count and treatment resistance (X 2 =9.53).
[0129] Kaplan-Meier analysis found that patients with low blast counts ( figure 2 ). These analyzes indicated that patient...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com