Circular nucleic acid vectors, and methods for making and using the same
A circular, vector technology, applied in the field of molecular biology, can solve the problems of slow recombination process, immunogenic integration mutation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
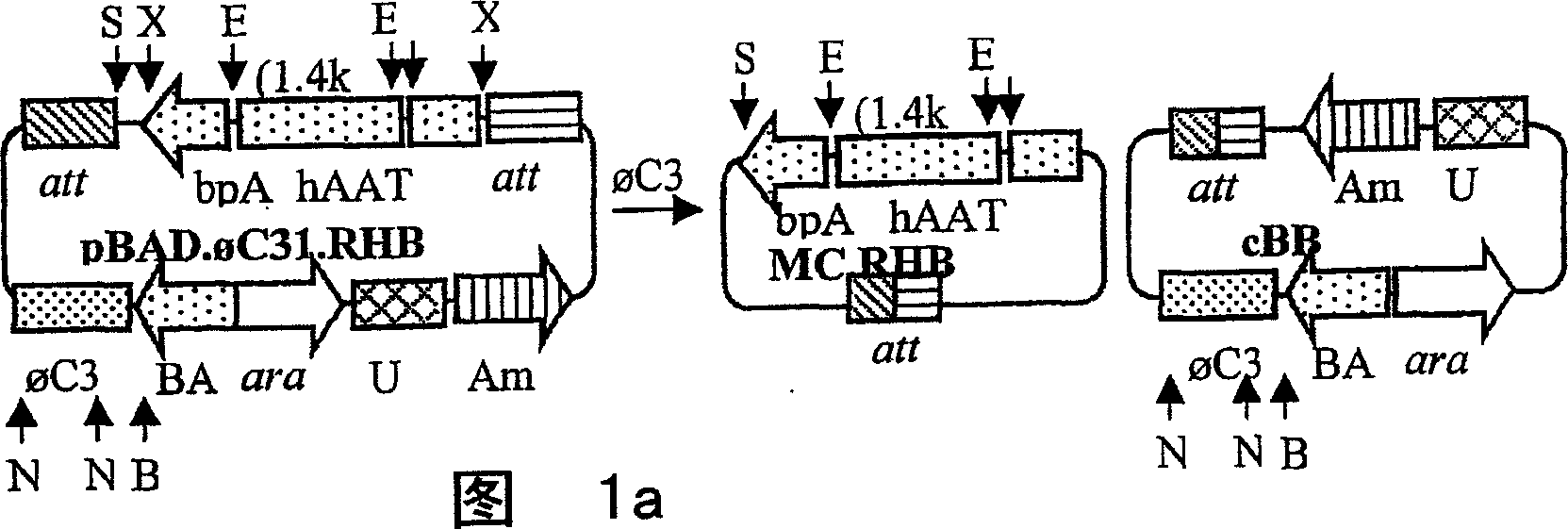
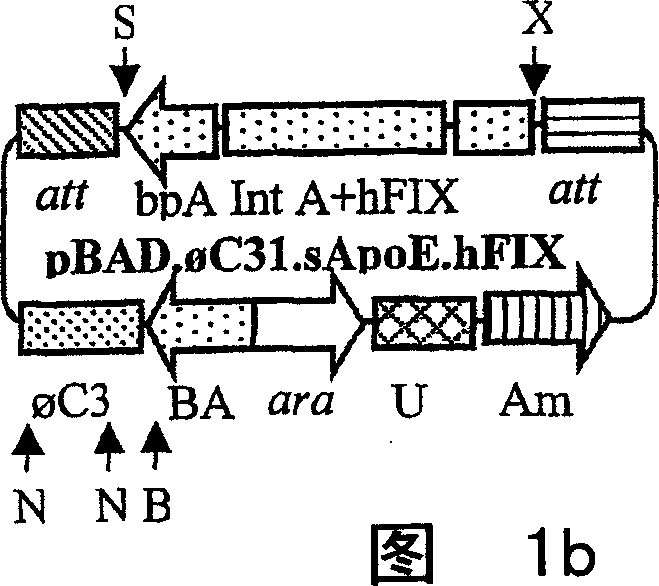
[0066] Preparation method of minicircle carrier
[0067] As outlined, the present invention also provides methods for efficiently preparing the minicircle vectors of the present invention. When the minicircle vector is prepared according to a specific embodiment of the present invention, the target expression cassette is contained, and the expression cassette is flanked by the parent nucleic acid of attB and attP sites and the unidirectional site-specific recombinase capable of recognizing attB and attP sites in a suitable Contacting under conditions, so that recombination is mediated by a unidirectional site-specific recombinase, so that the minicircle vector of the present invention is produced from the parental nucleic acid. "Both sides" refers to the expression cassette sequence, or other target sequences to be included in the minicircle vector, and each has an att site at its two ends, such as attB and attP. Therefore, the parental nucleic acid is represented by the follo...
Embodiment
[0082] I. Materials and methods
[0083] A. Construction of the carrier:
[0084]To make the hAAT minicircle construct pBAD.C31.RHB (Fig. 1a), we amplified the C31 integrase from pCMV.C31 (Groth, A.C., Olivares, E.C., Thyagarajan, B. & Calos, M.P. Aphage Integrase directs efficient site-specific integration in human cells.Proc NatlAcad SCI USA 97,5995-6000.(2000)), the primers used are: 5'-CCG TCC ATG GACACG TAC GCG GGT GCT (SEQ ID NO: 01) and 5′-ATG CGC GAG CTC GGT GTCTCG CTA CGC CGC TAC (SEQ ID NO: 02), the PCR product was inserted into the Nco I and Sac I sites of pBAD / Myc-His (Invitrogen, Carlsbad, CA) to obtain the intermediate plasmid pBAD .C31. We synthesized attB and attP using the corresponding DNA oligonucleotides (Groth et al., supra) and inserted them into the Spe I and Kpn I sites flanking the hAAT expression cassette on plasmid pRSV.hAAT.bpA, respectively (Fig. 2a). The attB, aatP binding sites and the expression cassette were inserted together between the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com