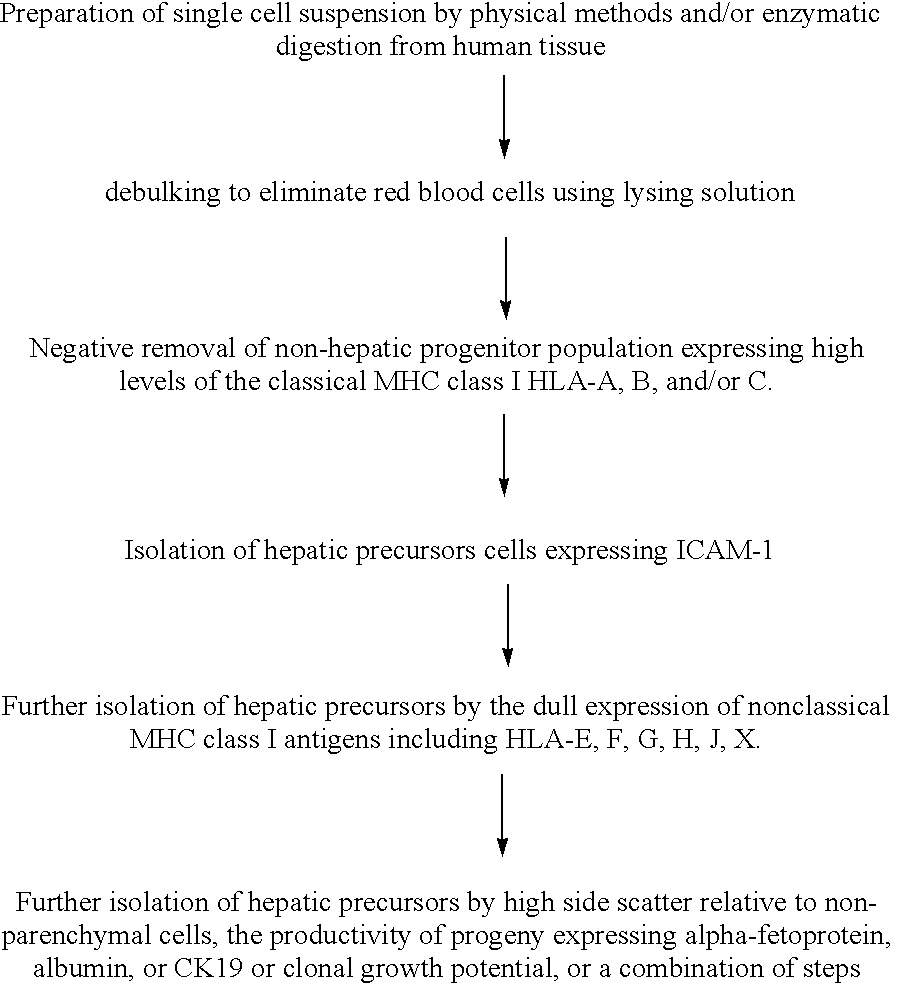
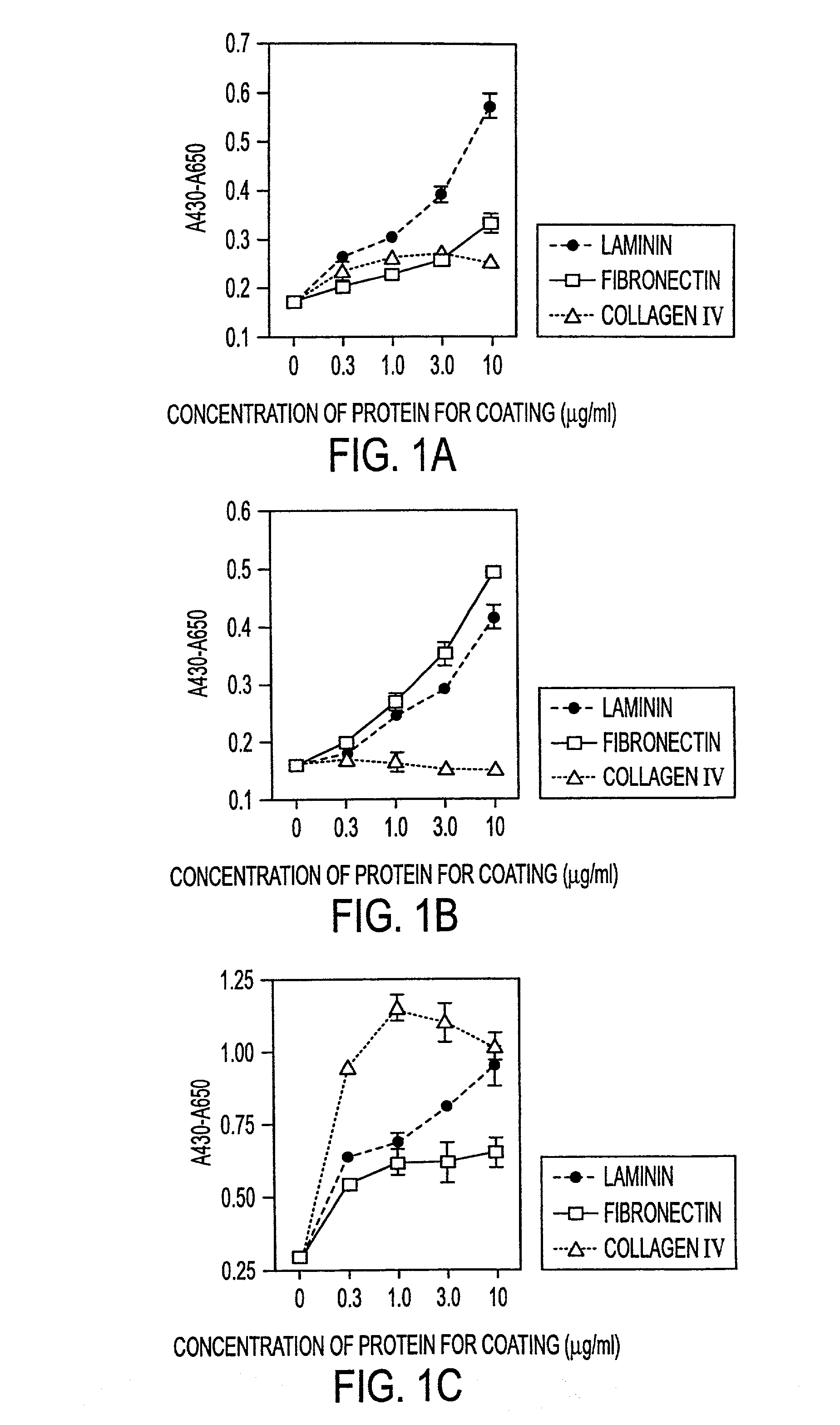
Methods of isolating bipotent hepatic progenitor cells
a technology of hepatic progenitor cells and methods, applied in biocide, instruments, genetic material ingredients, etc., can solve the problems of inability to test for bipotent cell populations, hematopoietic cells, and no evidence of biliary epithelial cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
Glossary
[0036] Classical MHC class I antigen. The group of major histocompatability antigens commonly found mostly on all nucleated cells although they are most highly expressed on hematopoietic cells. The antigen is also known as MDHC class Ia. The nomenclature of the classical MHC antigens is a function of species, for example in humans the MHC antigens are termed HLA. Table 3 provides nomenclature of classical MHC antigens in several species.
[0037] Non classical MHC class I antigen. The group of major histocompatability antigens, also known as MHC class Ib, that can vary even within a species. The nomenclature of the nonclassical MHC antigens varies by species, see, e.g., Table 4.
[0038] ICAM. Intercellular adhesion molecule-1(CD54) is a membrane glycoprotein and a member of the immunoglobulin superfamily. The ligands for ICAM-1 are the .beta.2-integrin, LFA-1 (CD11a / CD18) and Mac-1 (CD11b / CD18). This molecule is also important for leukocyte attachment to endothelium. In addition ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com