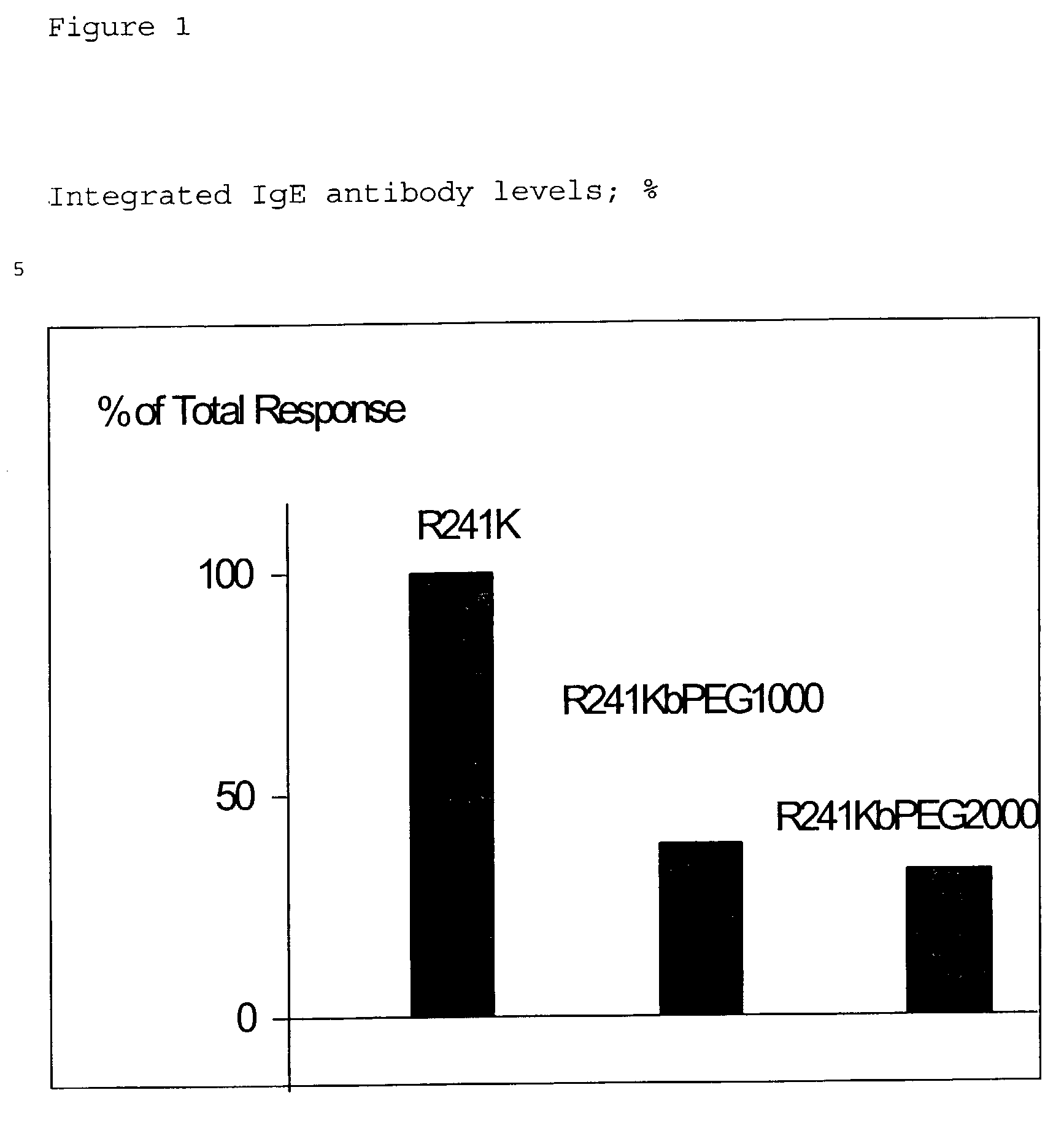
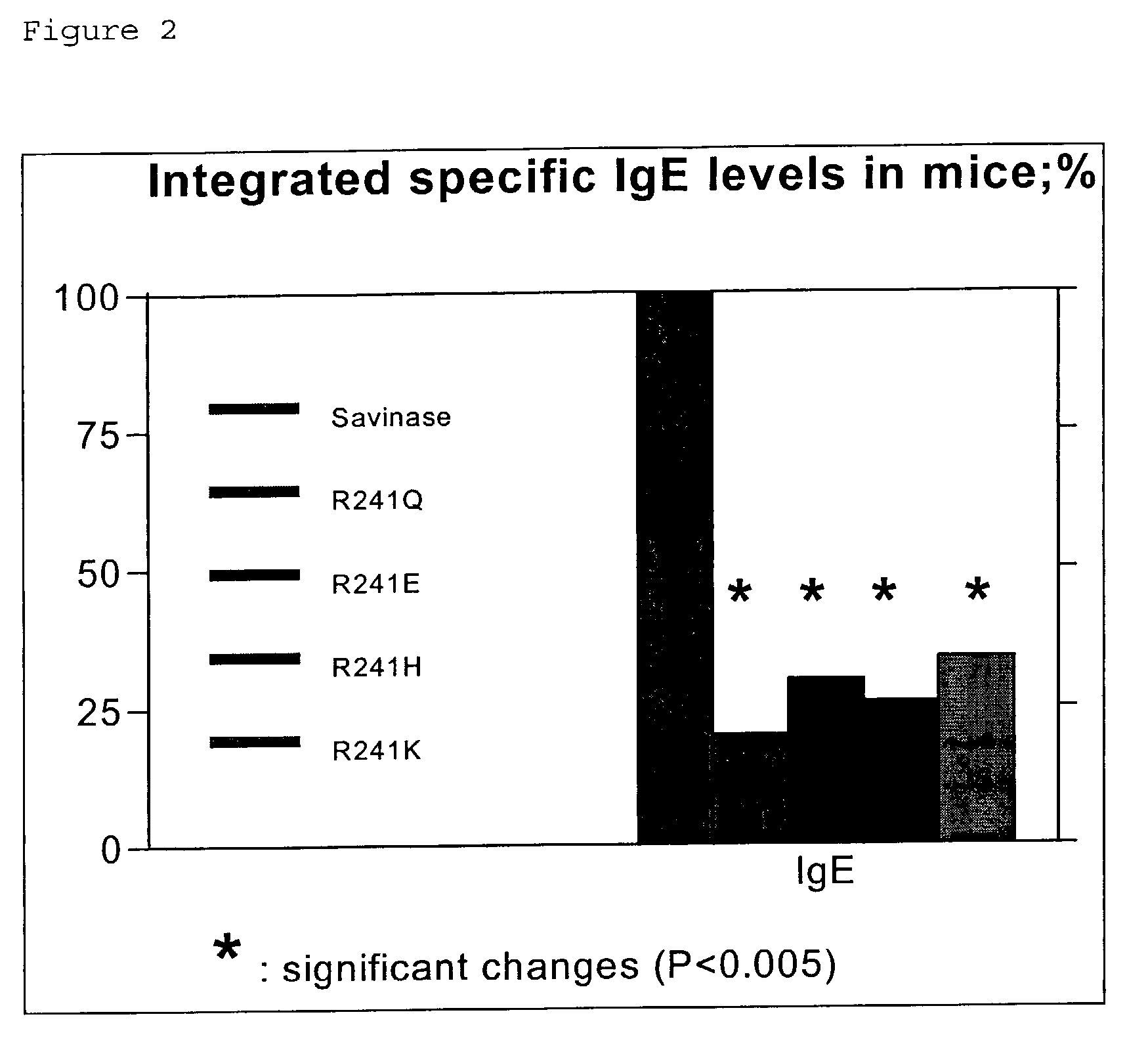
Modified polypeptides
a technology of polypeptides and polypeptides, applied in the field of modified polypeptides, can solve the problems of immunogenicity, potential risk of respiratory allergenicity, and immunogenicity of pharmaceutical polypeptides, and achieve the effect of reducing the risk of allergic reactions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0314] Subtilisin BPN'
[0315] In order to identify the residues to be modified, a distance and a directional criteria are applied.
[0316] As disclosed earlier residues having their C.sup.alpha-atom closer than 15 .ANG. to a ligand are targets for modification. Preferably, residues having their C.sup.beta-atom closer to the ligand bound than the C.sup.alpha-atom, thereby allowing a potential side chain to point in the direction of the ligand, are targets for modification.
[0317] The relevant distance can easily be measured using e.g. molecular graphics programs like InsightII from Molecular Simulations INC.
[0318] Especially surface exposed residues, defined as having ACC>0 when applying the DSSP program to the relevant protein part of the structure, are targets for modification. The DSSP program is disclosed in W. Kabsch and C. Sander, BIOPOLYMERS 22 (1983) pp. 2577-2637.
[0319] In Thomas E. Creighton, PROTEINS; Structure and Molecular Priciples, W H Freeman and Company, NY, ISBN: 0-7167...
example 2
[0325] PD498
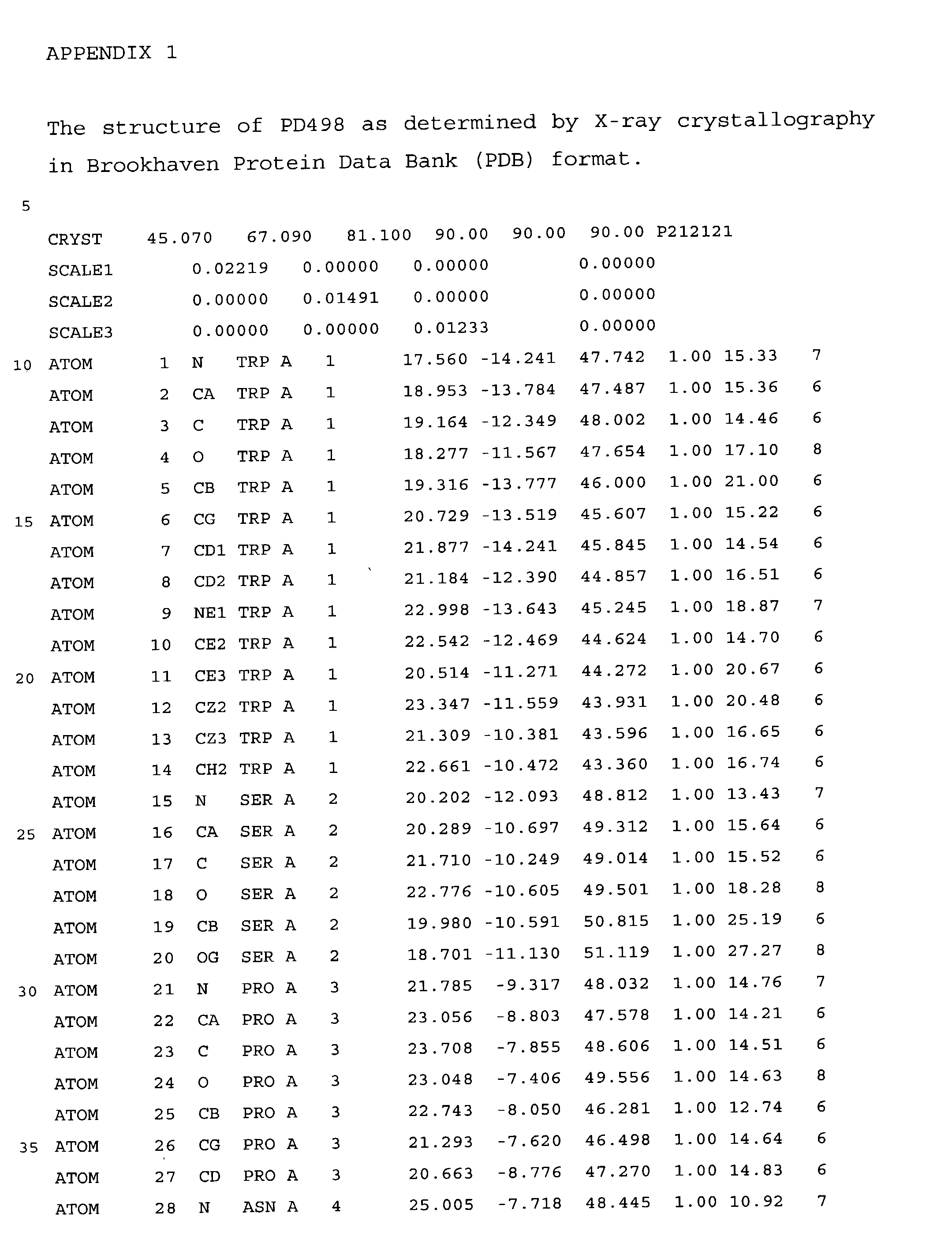
[0326] The 3-dimensional Structure of PD 498 as Determined by X-ray Crystallography in Brookhaven Protein Data Bank (PDB) Format
[0327] The sequence which was used to elucidate the three-dimensional structure forming the basis for the present invention consists of the 280 amino acids derived from Bacillus sp. PD498, NCIMB No. 40484 as disclosed in SEQ ID NO: 2.
[0328] The structure of PD498 was solved in accordance with the principle for X-ray crystallographic methods given in "X-Ray Structure Determination", Stout, G. K. and Jensen, L. H., John Wiley & Sons, inc. NY, 1989 and "Protein Crystallography" by Blundell, T. L. and Johnson, L. N., Academic Press, London, 1990. The structural coordinates for the solved crystal structure of PD 498 at 2.2 .ANG. resolution using the isomorphous replacement method are given in a standard PDB format (Brookhaven Protein Data Base) in Appendix 1. It is to be understood that Appendix 1 forms part of the present application.
[0329] In Appen...
example 3
[0333] Savinase
[0334] For this example the X-ray structure entry 1SVN in the Brookhaven Protein Data Bank was used. This structure contains two metal ions. Site 1 contains a calcium ion and is at a position equivalent to site 1 in subtilisin BPN'. Site 2 contains a calcium ion at a position equivalent to site 2 in subtilisin BPN'. In the following list a SEQUENTIAL numbering have been applied and NOT the numbering system used in the structure file.
6 Site 1: Resid resno dist(C.sup.alpha) dist(C.sup.beta) ACC (.ANG. .times. .ANG.) ACC (%) GLY 78 4.28 14 18.67 ASN 75 4.74 4.64 61 38.13 ASP 40 5.08 4.34 0 GLN 2 5.39 4.59 45 25.0 ALA 72 5.49 4.99 0 GLY 81 7.68 0 PRO 84 8.28 7.29 5 GLY 68 8.88 1 THR 202 9.19 8.67 0 HIS 38 10.40 9.89 13 PRO 5 10.47 10.26 14 9.66 ASN 42 10.55 10.50 94 58.75 TYR 208 10.72 9.76 65 28.26 GLN 200 11.75 11.39 82 45.56 ILE 8 12.10 10.58 3 PRO 14 12.91 12.63 49 33.79 THR 22 13.01 12.24 29 20.71 HIS 17 13.44 12.07 29 14.87 ALA 13 13.78 12.63 0 GLY 7 14.60 2 LEU 88 ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com