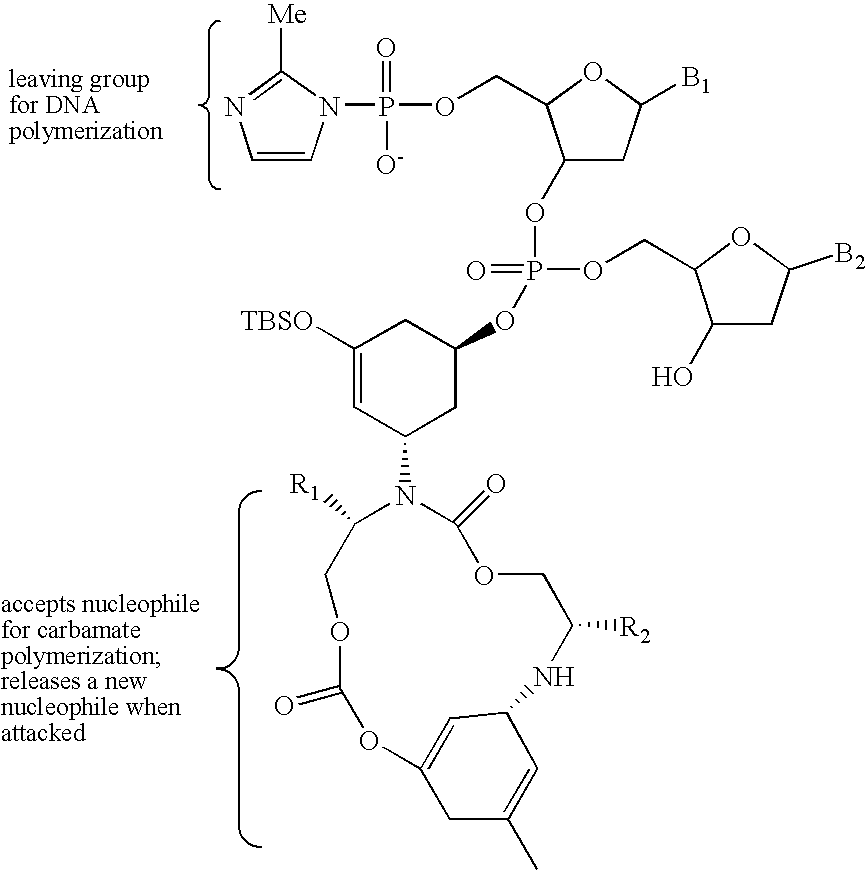
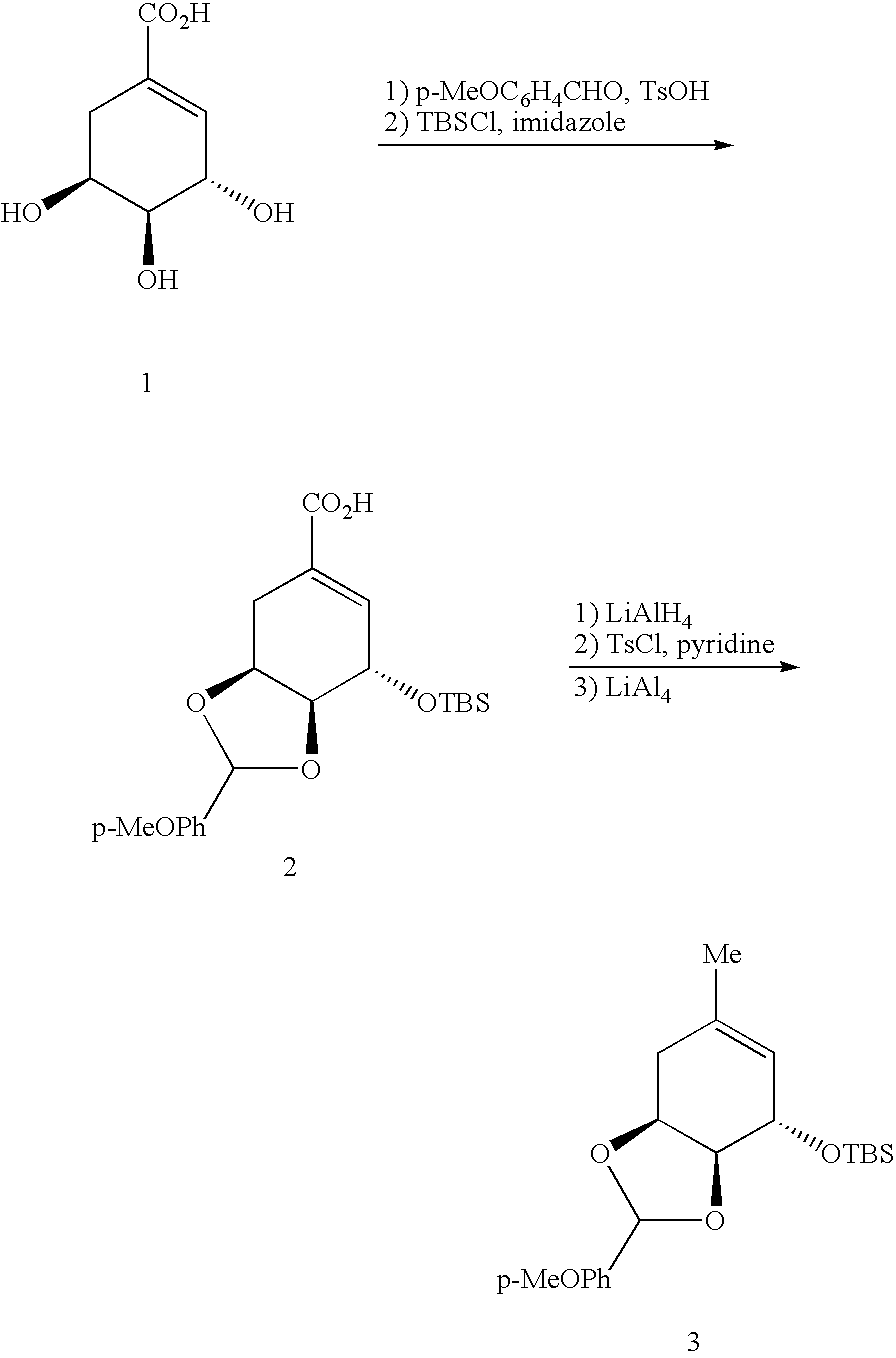
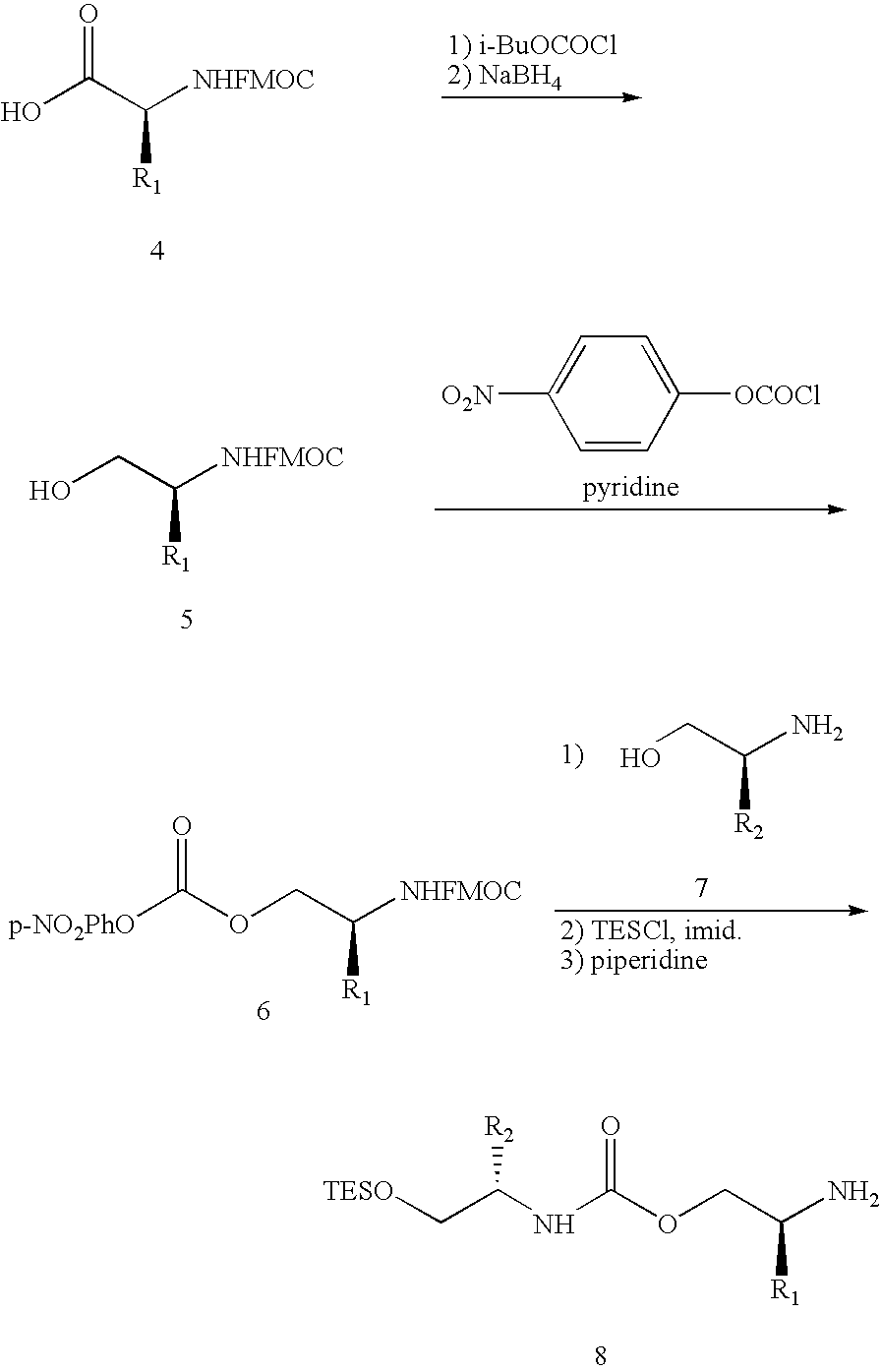
Evolving new molecular function
a new type of molecular function and molecular structure technology, applied in the field of evolution of new molecular function, can solve the problems of limiting the effectiveness of the chemical approach to generating molecular function, limiting the success of rational ligand or catalyst design, and inability to accurately predict the structural changes that will lead to new function, etc., to achieve a higher degree of activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
The Generality of DNA-Templated Synthesis
[0130] Clearly, implementing the small molecule evolution approach described above requires establishing the generality of DNA-templated synthesis. The present invention, for the first time, establishes the generality fo this approach and thus enables the syntheis of a vareity of chemical compounds using DNA-templated synthesis. As shown in FIG. 6a, the ability of two DNA architectures to support solution-phase DNA-templated synthesis was established. Both hairpin (H) and end-of-helix (E) templates bearing electrophilic maleimide groups reacted efficiently with one equivalent of thiol reagent linked to a complementary DNA oligonucleotide to yield the thioether product in minutes at 25° C. DNA-templated reaction rates (kapp=˜105 M−1s−1) were similar for H and E architectures despite significant differences in the relative orientation of their reactive groups. In contrast, no product was observed when using reagents containing sequence mismatc...
example 2
Exemplary Reactions for Use in DNA-Templated Synthesis
[0145] As discussed above, the generality of DNA-templated synthetic chemistry was examined (see, Liu et al. J. Am. Chem. Soc. 2001, 123, 6961). Specifically, the ability of DNA-templated synthesis to direct a modest collection of chemical reactions without requiring the precise alignment of reactive groups into DNA-like conformations was demonstrated. Indeed, the distance independence and sequence fidelity of DNA-templated synthesis allowed the simultaneous, one-pot translation of a model library of more than 1,000 templates into the corresponding thioether products, one of which was enriched by in vitro selection for binding to the protein streptavidin and amplified by PCR.
[0146] As described in detail herein, the generality of DNA-templated synthesis has been further expanded and it has been demonstrated that a variety of chemical reactions can be utilized for the construction of small molecules and in particular, for the fi...
example 3
Development of Exemplary Linkers
[0158] As will be appreciated by one of ordinary skill in the art, it is frequently useful to leave the DNA moiety of the reagents linked to products during reaction development to facilitate analysis by gel electrophoresis. The use of DNA-templated synthesis to translate libraries of DNA into corresponding libraries of synthetic small molecules suitable for in vitro selection, however, requires the development of cleavable linkers connecting reactive groups of reagents with their decoding DNA oligonucleotides. As described below and herein, three exemplary types of linkers have been developed (see, FIG. 18). For reagents with one reactive group, it would be desirable to position DNA as a leaving group to the reactive moiety. Under this “autocleavable” linker strategy, the DNA-reactive group bond is cleaved as a natural consequence of the reaction. As but one example of this approach, a fluorescent Wittig phosphorane reagent (14, referring to FIG. 19...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com