Methods to identify compounds useful for the treatment of proliferative and differentiative disorders
a technology of proliferative and differentiative disorders and compounds, applied in the direction of biological material analysis, drug compositions, cardiovascular disorders, etc., can solve the problems of abnormal degradation of negative regulators (tumor suppressor proteins), cell proliferation too quickly, cancer,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
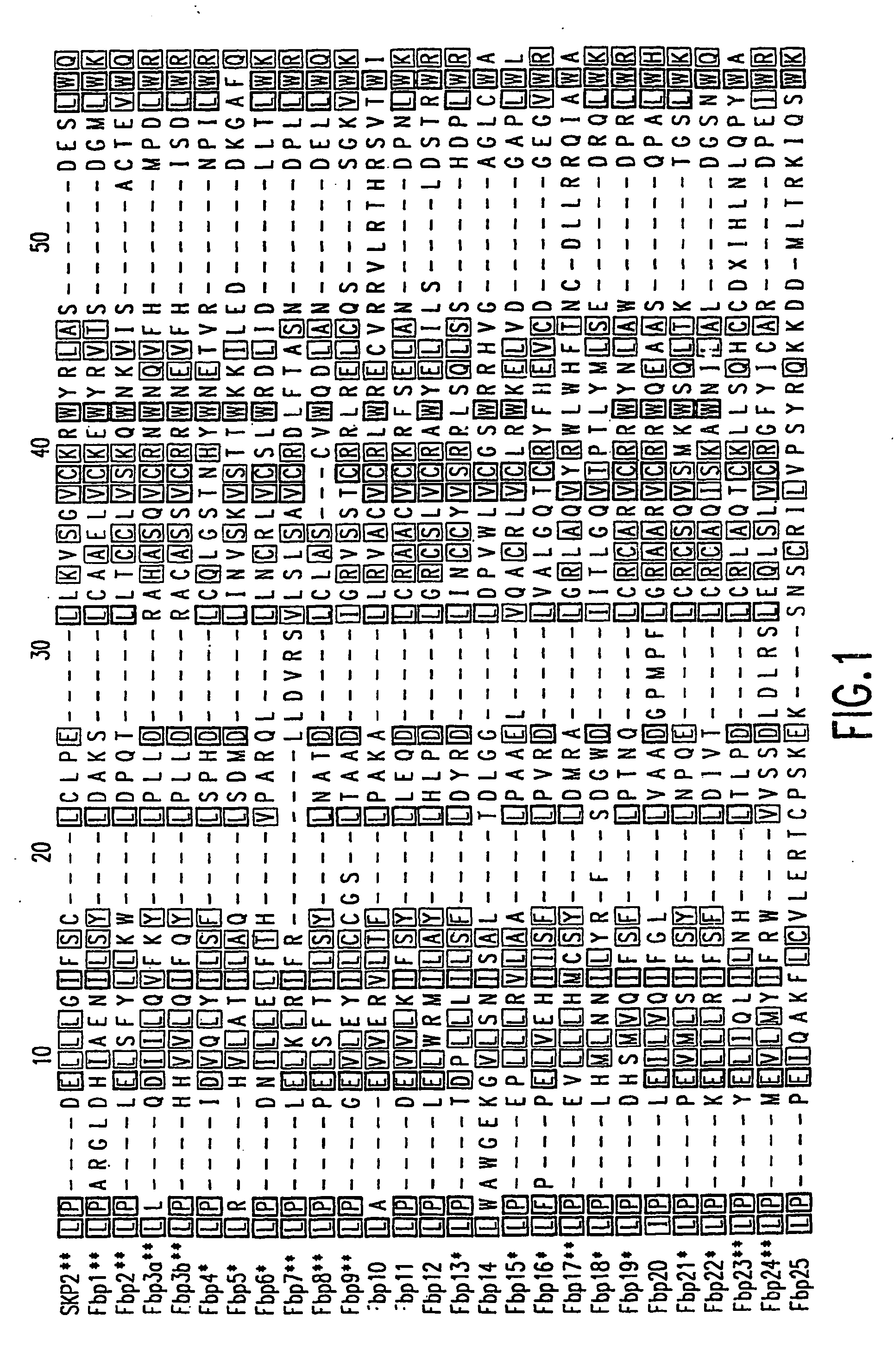
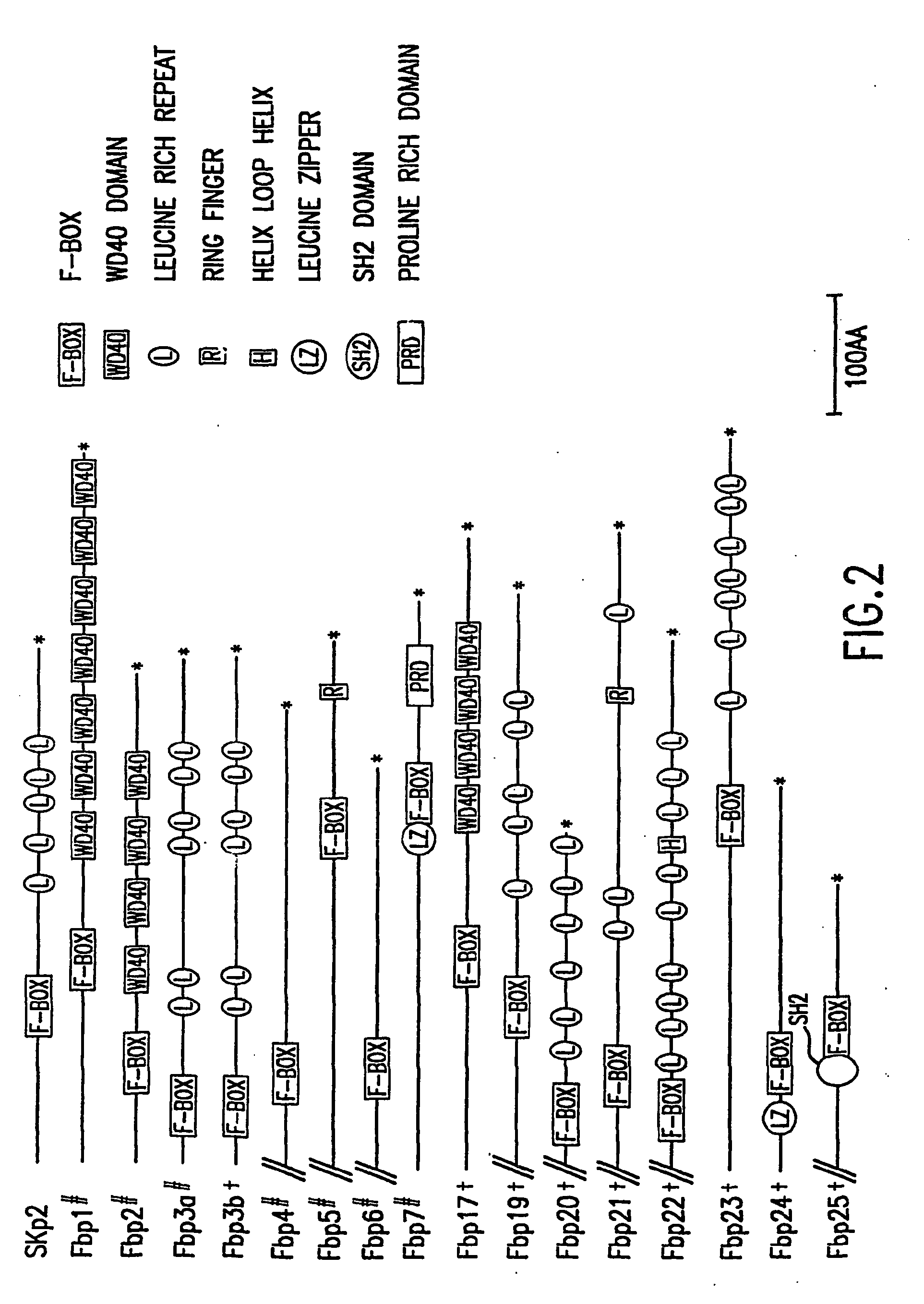
[0129] The present invention relates to novel F-box proteins and to novel substrates of F-box proteins. The present invention relates to screening assays designed to identify substrates of the F-box proteins and to identify small molecules and compounds which modulate the interaction and / or activity of the F-box proteins and their substrates.
[0130] The present invention relates to screening assays to identify substrates of the novel F-box proteins and to identify potential therapeutic agents. The present invention further relates to screening assays based on the identification of novel substrates of both novel and known F-box proteins. The screening assays of the present invention may be used to identify potential therapeutic agents which may be used in protocols and as pharmaceutical compositions designed to target the novel ubiquitin ligases and interactions with their substrates for the treatment of proliferative disorders. In one particular embodiment the present invention rela...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com