High throughput detection of molecular markers based on aflp and high through-put sequencing
a molecular marker and high throughput technology, applied in the field of molecular biology and biotechnology, can solve the problems of high throughput of aflp, and insufficient etc., to achieve efficient and reliable improvement, adequate identification of restriction fragments, and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
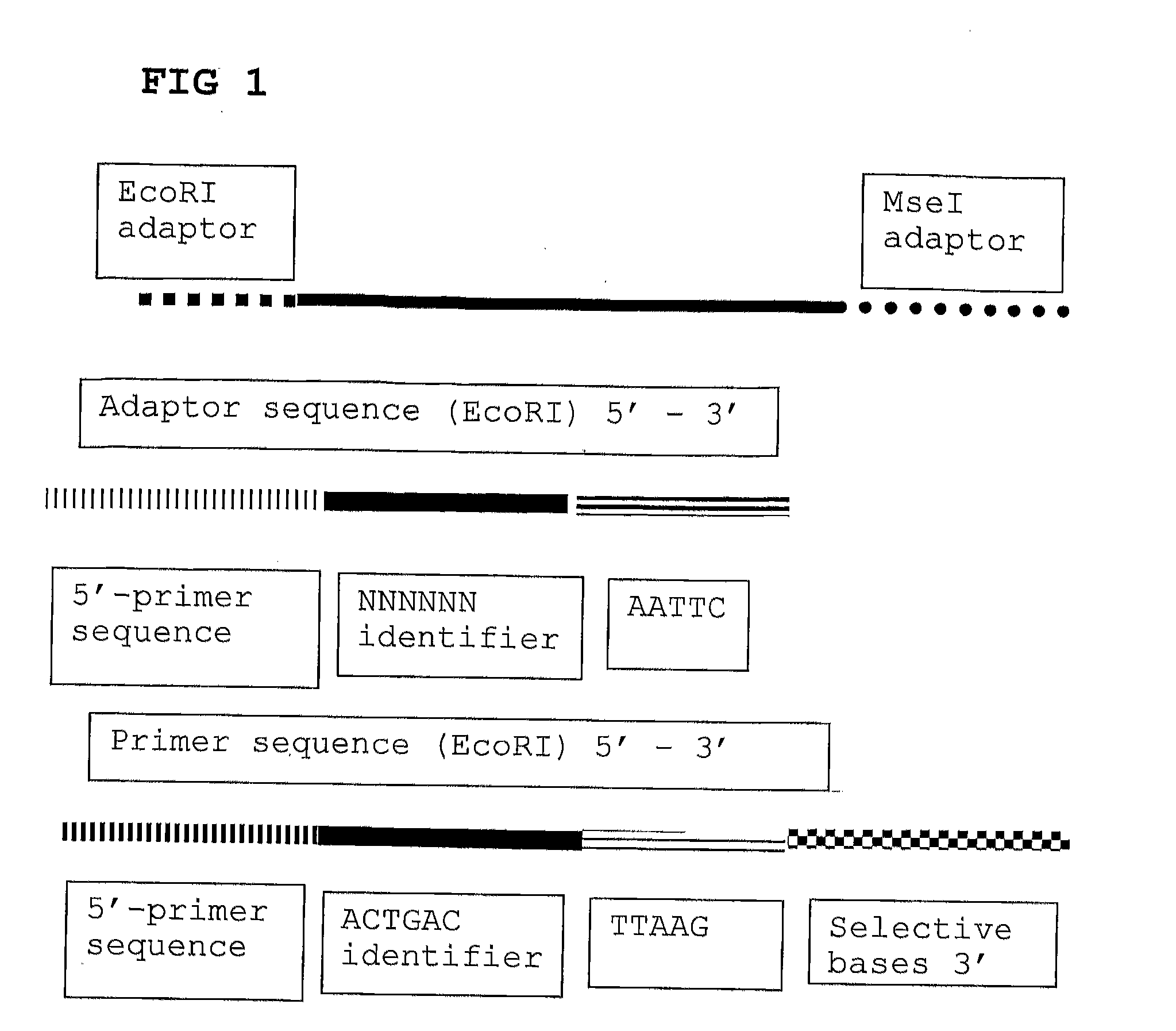
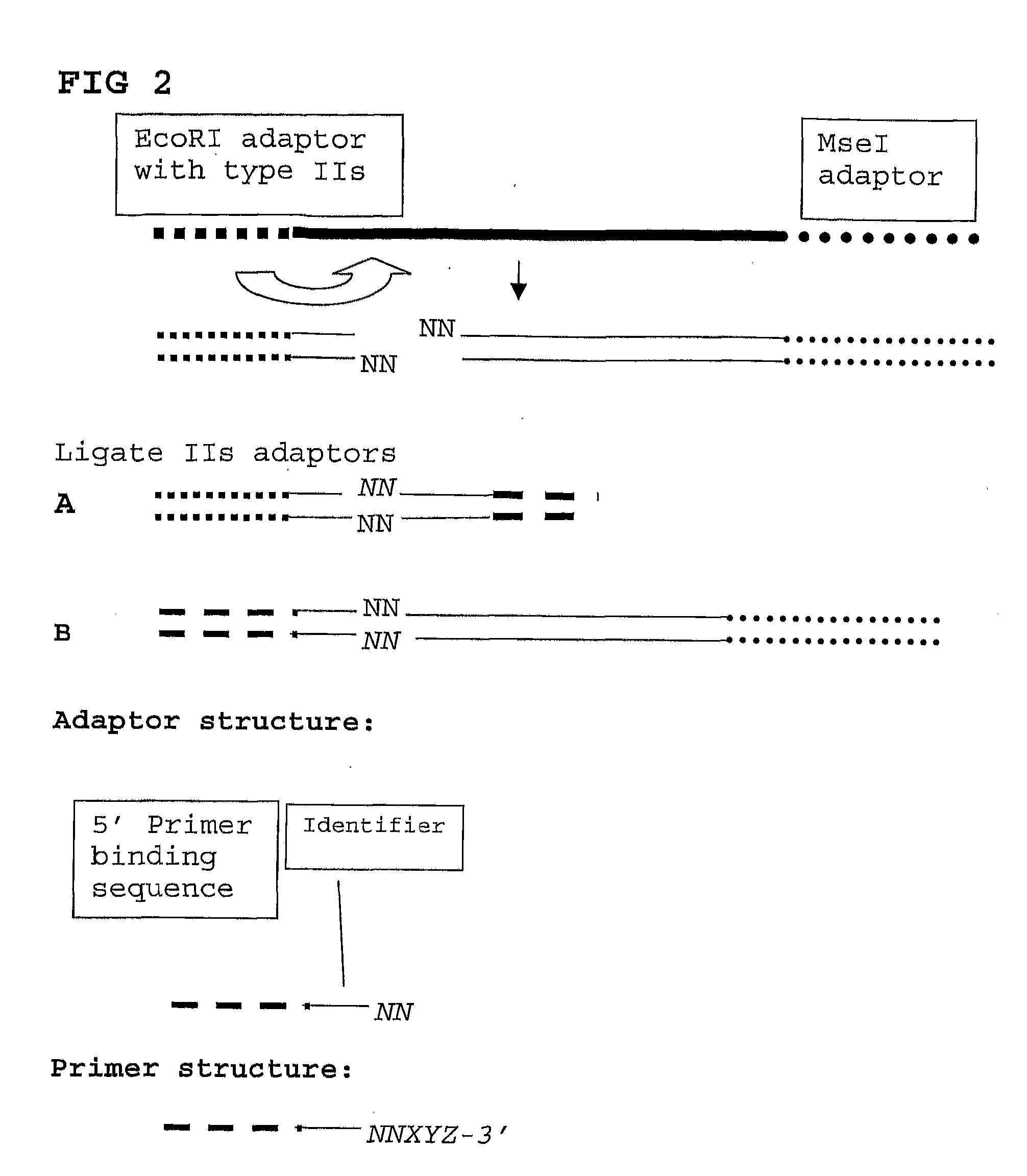
[0093]For each DNA sample a restriction-ligation step is performed using EcoRI and MseI as enzymes. Adaptors are based on the hybridizing sequences located on the surface of the Solexa high throughput sequencing system, more in particular the EcoRI adapter contains the P5 sequence (sequence primer part) and the MseI adaptor contains the P7 sequence (bridge PCR primer sequence). The EcoRI adaptor further contains the sample identifying tag. 96 different EcoRI adaptors and one MseI adaptor are used. It is possible to use a degenerated EcoRI adaptor. The template preparation is inclusive of a size selection step by incubation of the mixture for 10 minutes at 80 degrees Celsius after the restriction (EcoRI+MseI) step but prior to the adapter ligation step. Fragments smaller than 130 nt are removed (in a maize sample).
[0094]The complexity of the mixture is reduced by a selective preamplification using +1 primers (i.e. containing one randomly selective nucleotide at the 3′ end, using 96 E...
example 2
[0097]Sequence-based detection of AFLP fragments was performed using Solexa's Clonal Single Molecule Array (CSMATM) technology, a Sequencing-by-Synthesis platform capable of analyzing up to 40 million individual fragments in a single sequence run.
[0098]The experimental sequence involves AFLP template preparation, selective (AFLP) amplification, single molecule bridge amplification and sequencing of millions of sequence tags from one restriction enzyme end of the AFLP fragments. Maize parental lines B73 and Mo17 and 87 Recombinant Inbred Lines (RILs) were used and sequenced over 8.9 million EcoRI AFLP fragment termini were sequenced to provide proof-of-principle for sequence-based AFLP detection.
[0099]Parental lines B73 and Mo17 and 87 RILs were selected. AFLP templates were prepared using restriction enzyme combination EcoRI / MseI. Selective amplification was performed using +21+3 AFLP primers.
[0100]Template fragments for Solexa CSMA bridge amplification were prepared by performing a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| genome size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com