Carbapenemases for use with antibiotics for the protection of the intestinal microbiome
a technology of antibiotics and carbapenemases, which is applied in the direction of antibacterial agents, drug compositions, peptide/protein ingredients, etc., can solve the problems of further disease, serious illnesses, and disruption of the microbiome, and achieve the effect of protecting the microbiom
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Design of P2A, NDM-1, and KPC-1 / 2 E. coli Expression Plasmids and Generation of Transformed Bacterial Strains
[0232]The purpose of this study is, among others, to generate a panel of transformed bacterial strains to screen for carbapenemase expression.
[0233]For E. coli-mediated expression of the carbapenemases, P2A, NDM-1 (Yong et al., Antimicrob. Agents Chemother. (2009), 53:5046-5054) and KPC1 / 2 [Yigit et al., Antimicrob. Agents Chemother. (2001) 45:1151-1161,Yigit et al., Antimicrob. Agents Chemother. (2003), 47:3881-3889)], a total of 39 expression plasmids and 104 bacterial strains were generated. For P2A, 3 gene variants (SEQ ID NOS: 1-3), 9 plasmids and 25 bacterial strains, for NMD-1, 8 gene variants (SEQ ID NOS: 4-11), 17 plasmids and 44 bacterial strains, and for KPC1 / 2, 7 gene variants (SEQ ID NOS: 12-18), 13 plasmids and 35 bacterial strains were generated and tested. The gene expression constructs differed by plasmid backbone (e.g., pBR322 or pUC, for medium and high cop...
example 2
Screening Various P2A, NDM, and KPC-Encoding Bacterial Strains
[0237]Experiments were carried out, inter alia, to identify bacterial strains that result in sufficient expression of one or more biologically active carbapenemases, among other purposes.
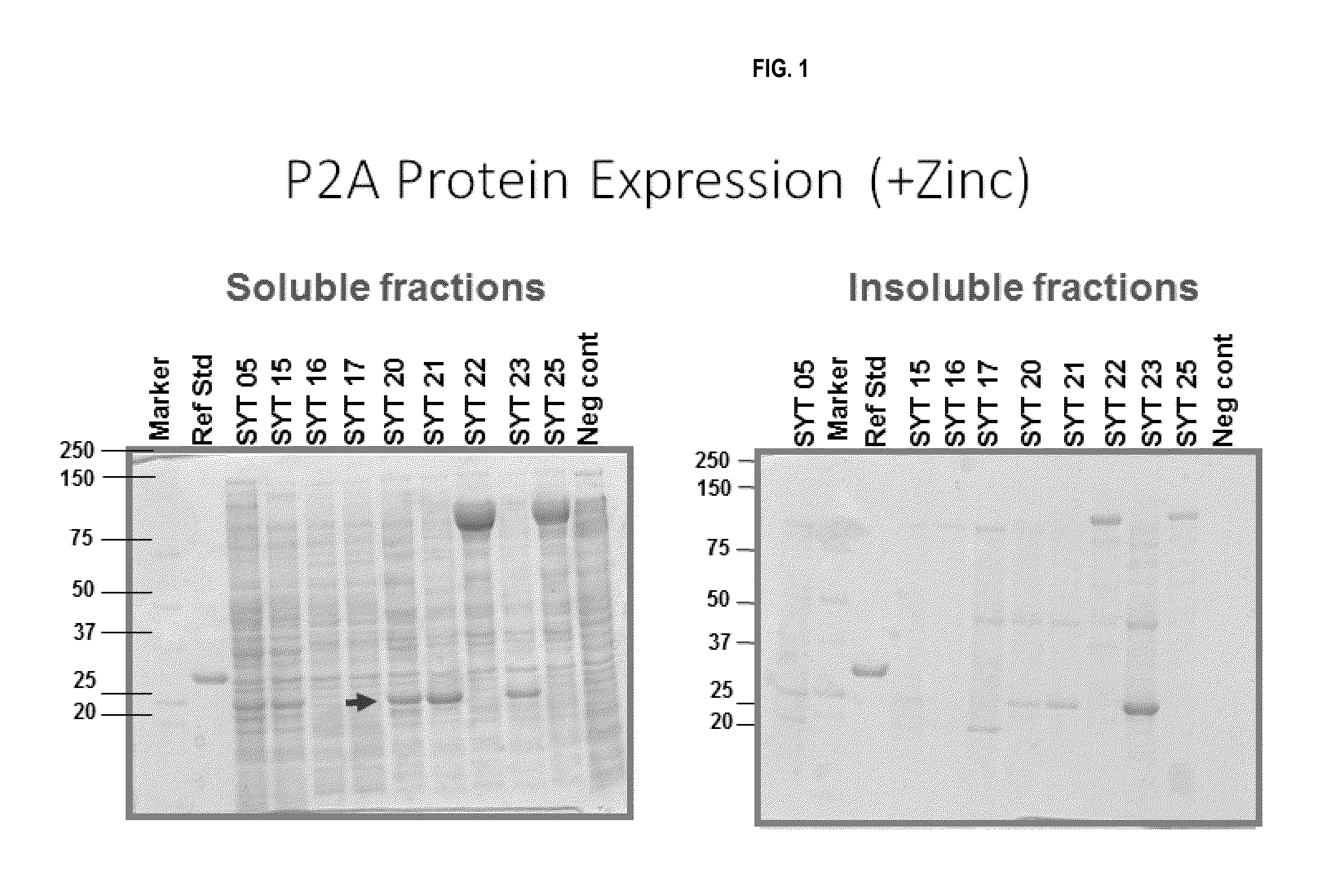
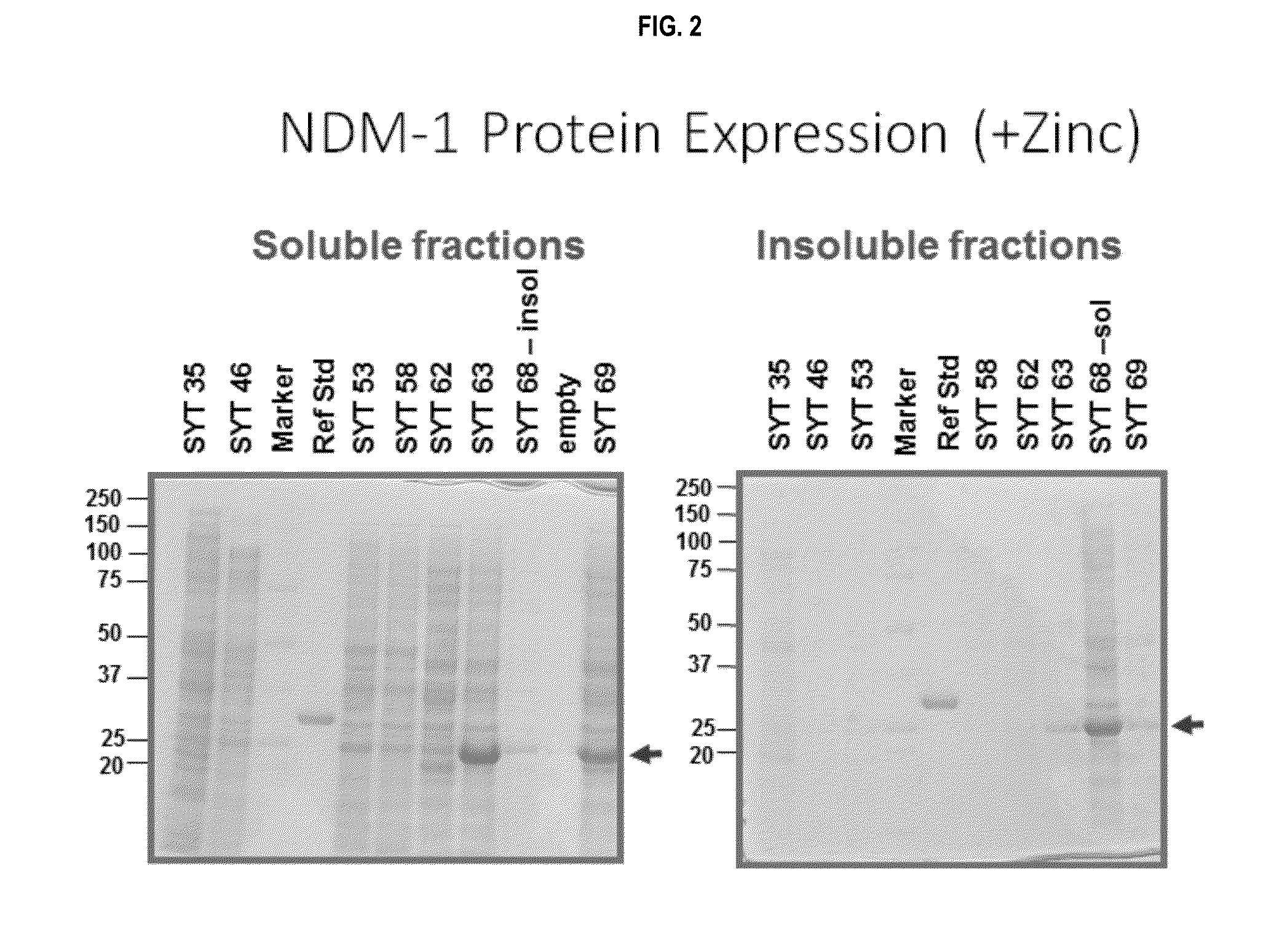
[0238]A total of 104 transformed bacterial strains, 25 P2A strains, 44 NDM strains, and 35 KPC strains (as shown in Tables 1-3) were assessed for carbapenemase protein expression in a screening assay. Colonies of the 104 transformed strains were picked, grown overnight, diluted 1:50 into 24 well dishes (3 ml volume per well), and grown overnight. A total of 2×1.0 ml sample were saved per strain. Bacteria were lysed with BugBuster protein extraction reagent (EMD Millipore, Cat #70584). The soluble and insoluble fractions were analyzed using SDS-PAGE. The data from the initial screen of P2A and NDM bacterial strains is summarized in Tables 5 and 6. 8 out of 25 P2A strains produced a soluble or insoluble band (Table 5), and 14 out of 44 NMD ...
example 3
Evaluation of Enzyme Expression Levels, Biological Activity, and Reproducibility
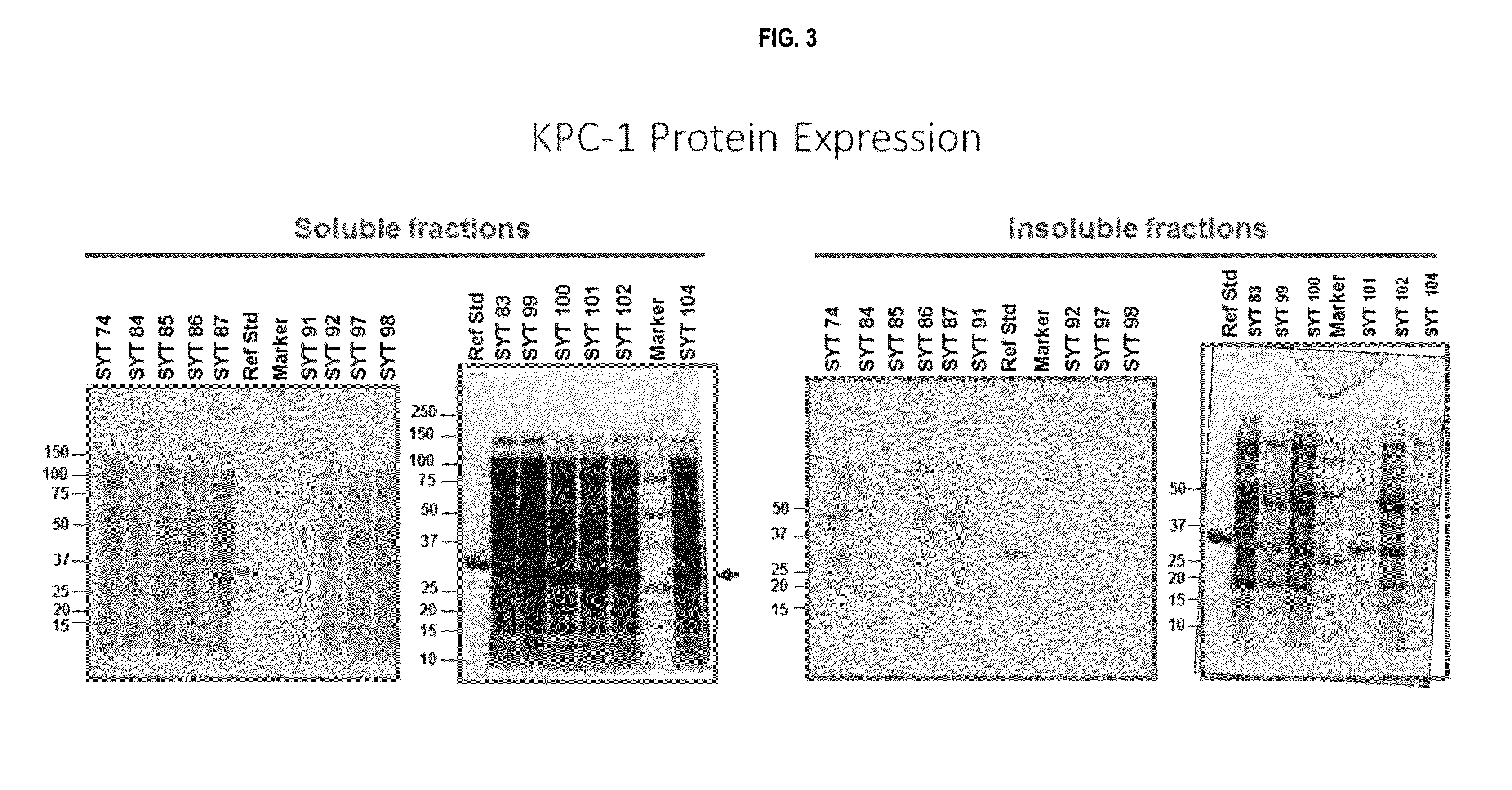
[0244]Experiments were carried out, inter alia, to evaluate the growth and enzyme production characteristics of the E. coli carbapenemase-expressing strains when scaled up from 24-well plates into shake flasks.
[0245]The strains chosen for scale-up into shake flasks were P2A-21, NDM-63, NDM-68, and NDM-69, and KPC-101 and KPC-102, based on protein expression levels and biological activities. All strains were streaked onto LB agar plates containing tetracycline (tet) (12.5 ug / ml) and incubated at 30° C. for approximately 18 hours. For each strain, an isolated colony was used to inoculate a 2 ml pre-culture in LB tet broth (12.5ug / ml) and incubated at 30° C. overnight. The pre-culture was inoculated 1 / 50 into 50 ml LB tet (12.5 ug / ml) supplemented with 100 uM ZnSO4 in two, 500 ml baffled flasks with ventilated caps and incubated at 30° C. with shaking for approximately 24 hours. 1 ml aliquots were centrifug...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com