Method of isolating nucleic acids for long sequencing reads
a technology of nucleic acids and reads, applied in the field of nucleic acid isolation, can solve the problems of inability to recover short strands, inability to detect long strands, etc., and achieve the effect of high ionic strength
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
HIV-1 RNA from Human Plasma Samples
Clinical Specimens:
[0027]Plasma samples of six patients, including HIV Subtype A and B, were obtained. The samples were quantified for viral loads and genotyped using COBAS® Ampliprep / COBAS® TaqMan® (CAP / CTM) platform (Roche Diagnostics, GmbH, Mannheim, Germany). Samples with a viral load of about 100,000 copies / ml were selected. The samples were diluted in HIV negative plasma to the concentrations used in this study.
Viral RNA Isolation:
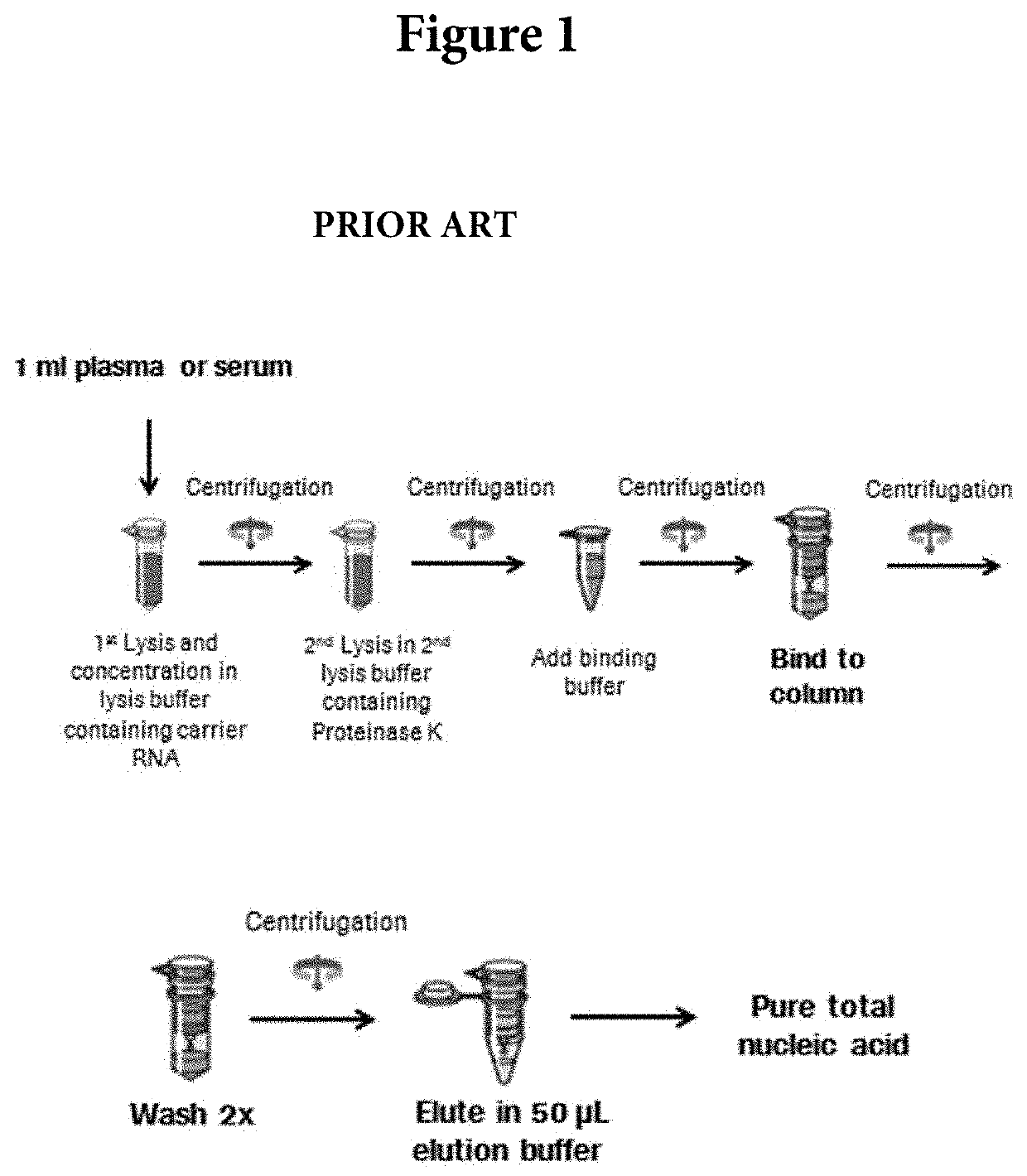
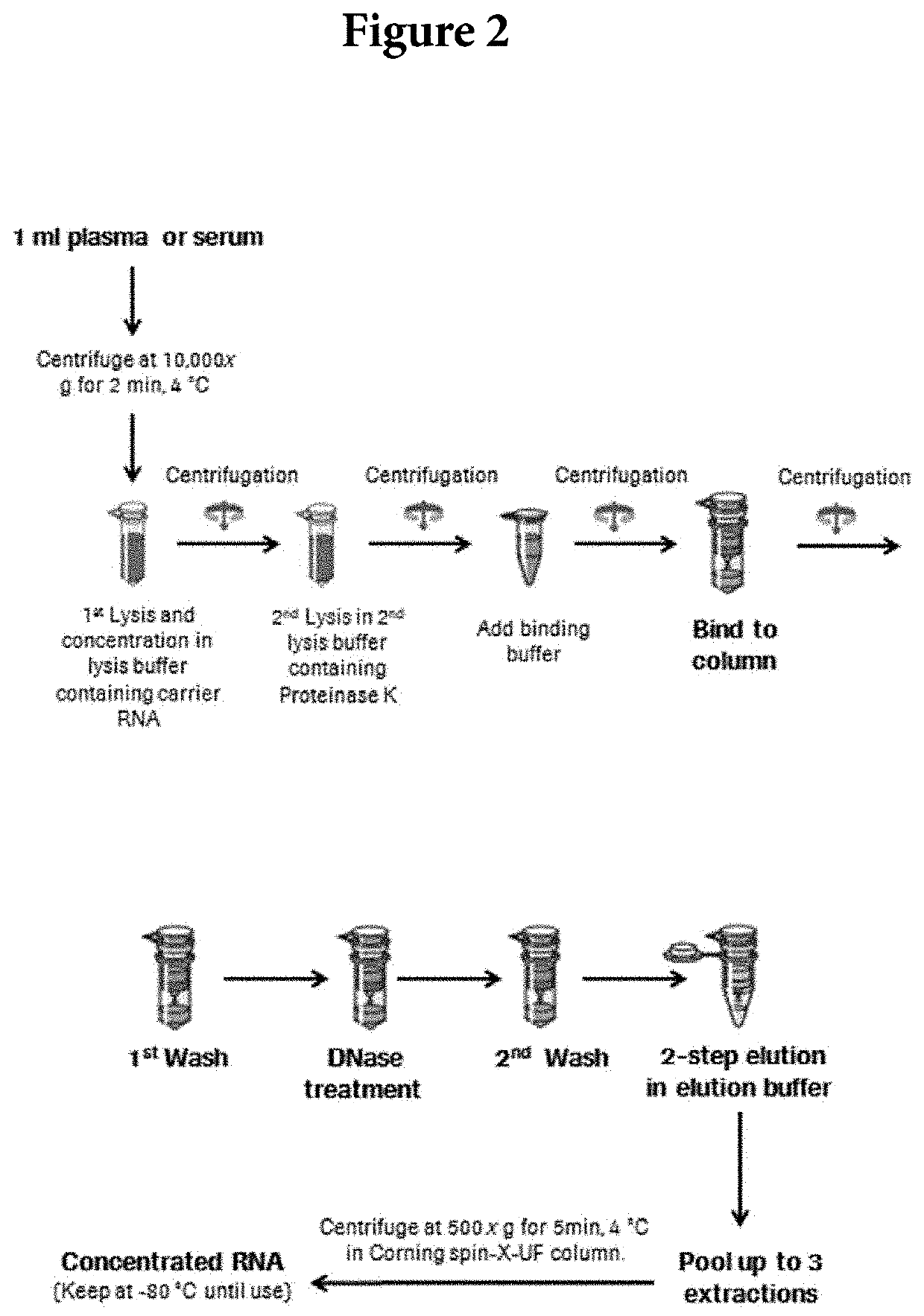
[0028]The viral RNA isolation in this study was performed using the QIAamp UltraSens Virus Kit from QIAgen (Valencia, Calif.). The default method was performed per the manufacturer's protocol with input volume of 1.0 ml plasma sample and final elution volume at 50 μL. For the in-house optimized protocol, approximately 1.2 ml of plasma samples was centrifuged at 10,000×g at 4° C. for 2 minutes. The isolation was performed per manufacturer's instructions with the following modifications. After the first wash step, 30U...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com