Probe and primer for detecting novel coronavirus SARS-CoV-2, kit, detection method and application
A sars-cov-2, sars-cov-2n technology, applied in the field of probes and primers for the detection of new coronavirus SARS-CoV-2, can solve problems such as increasing the risk of new coronavirus infection, avoid cross-transmission, control Virus outbreak, time saving effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
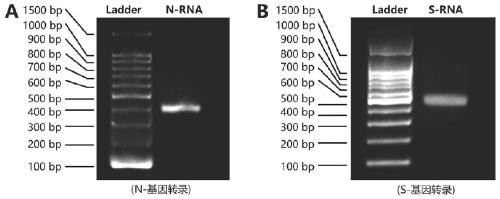
[0066] Example 1: Using T7 transcriptase to prepare N and S gene fragments of SARS-CoV-2 new coronavirus as a positive control.
[0067] Submit the S and N gene sequences of the new coronavirus to Langking Biotechnology (Shanghai, China) for gene synthesis. The synthetic new coronavirus N gene sequence is:
[0068] CATATGTCTGATAATGGACCCCAAAATCAGCGAAATGCACCCCGCATTACGTTTGGTGGACCCTCAGATTCAACT GGCAGTAACCAGAATGGAGAACGCAGTGGGGCGCGATCAAAACAACGTCGGCCCCAAGGTTTACCCAATAATAC TGCGTCTTGGTTCACCGCTCTCACTCAACATGGCAAGGAAGACCTTAAATTCCCTCGAGGACAAGGCGTTCCAA TTAACACCAATAGCAGTCCAGATGACCAAATTGGCTACTACCGAAGAGCTACCAGACGAATTCGTGGTGGTGAC GGTAAAATGTAAAAGCTT。
[0069] The synthetic new coronavirus S gene sequence is:
[0070] CATATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGTGTTAAATCTTACAACCAGAACTCAATTACCCCCTGCATACACTAATTCTTTCACACGTGGTGTTTTATTACCCTGACAAAGTTTTTCAGATCCTCAGTTTTCATTCAACTCAGACTTGTTCTTACCTTTCTTTTCCAATGTTACTTGGTTCCATGCTATACATGTCCTCTGGGACCAGAGGGTAAACCTA.
[0071] The above N gene and S gene were r...
Embodiment 2
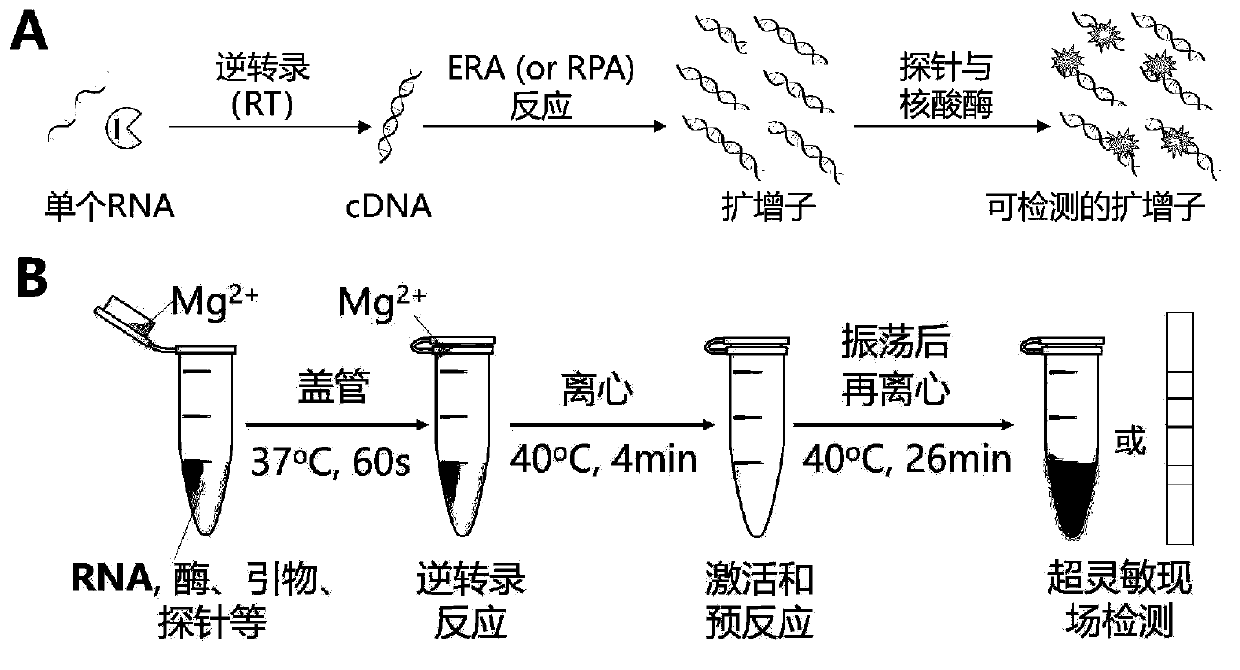
[0074] Embodiment 2. Based on the WEPEAR method, use the exo probe to detect the N gene of the new coronavirus SARS-CoV-2 nucleic acid RNA.
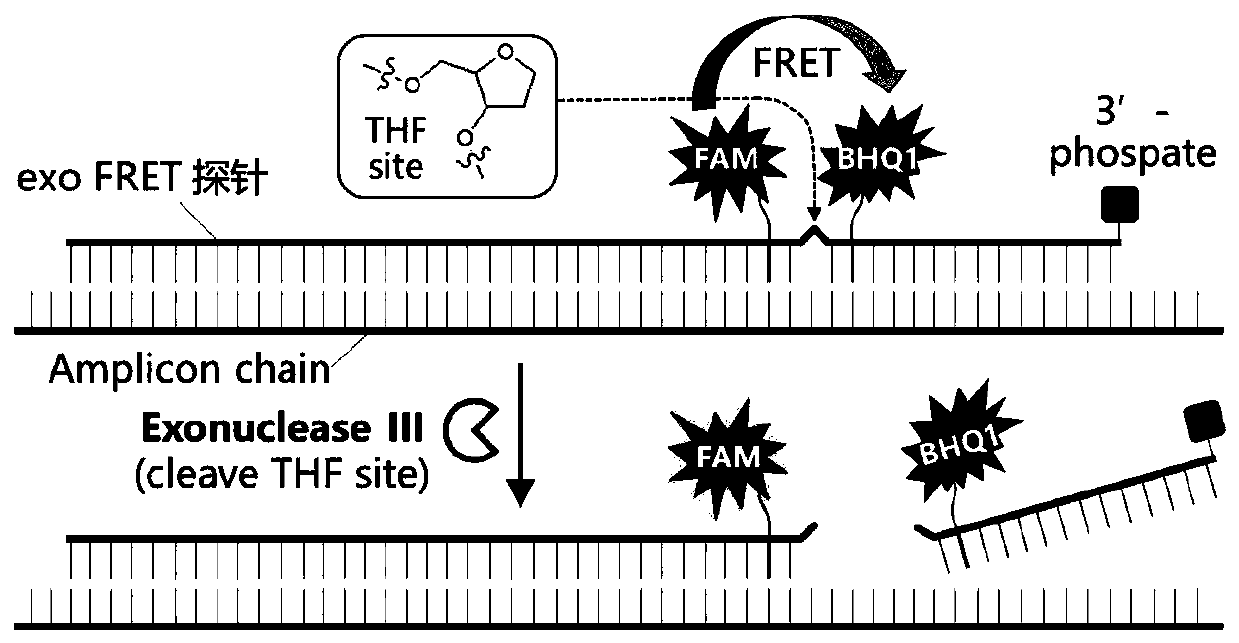
[0075] Regarding the detection of nucleic acid amplification products by using exo probes, it is necessary to design forward primers, reverse primers, and exo probes. The design and working principle of exo probes are as follows image 3 shown. The forward and reverse primers are about 33bp in length (30-35bp), and can amplify fragments with a length of 100-200bp. The characteristic of the exo probe is that there is an alkali-free tetrahydrofuran (THF) residue (idSp) in the middle of the probe, which can be cut by exonuclease III enzyme under appropriate conditions, and there is a base separated from the 5'-side of the THF residue There is a 6-FAM fluorescein-modified thymine T-residue (i6FAMdT), separated by one base on the 3'-side of the THF residue. There is a BHQ1 black hole quencher-modified thymine T-residue (iBHQ1dT ), so there ...
Embodiment 3
[0083] Example 3: Using exo probes to amplify based on the WEPEAR method and RT-ERA Real-time fluorescent quantitative PCR instrument is used to detect the N gene of SARS-CoV-2 in real time.
[0084] In this embodiment, eight tubes with a volume of 250 μl with a transparent flat cover are used for the reaction, and various parameters need to be correctly set in the use of the real-time fluorescent quantitative PCR instrument. The primers, probes and reagents used in the detection process of this example are basically the same as those in Example 2, but the specific operation process has certain changes.
[0085] Specifically, first take an eight-strip tube with a transparent flat cover, add 1 μl of the nucleic acid sample to be tested to the bottom of the eight-strip tube, and at the same time add 2 μl of Mg(OAc) 2 The activator is added to the cap of the eight-tube tube. For a 50 μl reaction system, 2.5 μl of exo forward primer (10 μM), 2.5 μl of exo reverse primer (10 μM), ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com