New micro ribonucleic acid quantitative PCR (polymerase chain reaction) detection method
A technology for target nucleic acid and quantitative analysis, applied in the field of novel microRNA quantitative PCR (polymerase chain reaction) detection, which can solve the problem that TaqMan-MGB fluorescent probe is expensive to synthesize, fails to detect mature microRNAs, and is difficult to detect. Detection methods and other problems, to avoid the formation of dimers, solve the huge consumption of samples, and improve the specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
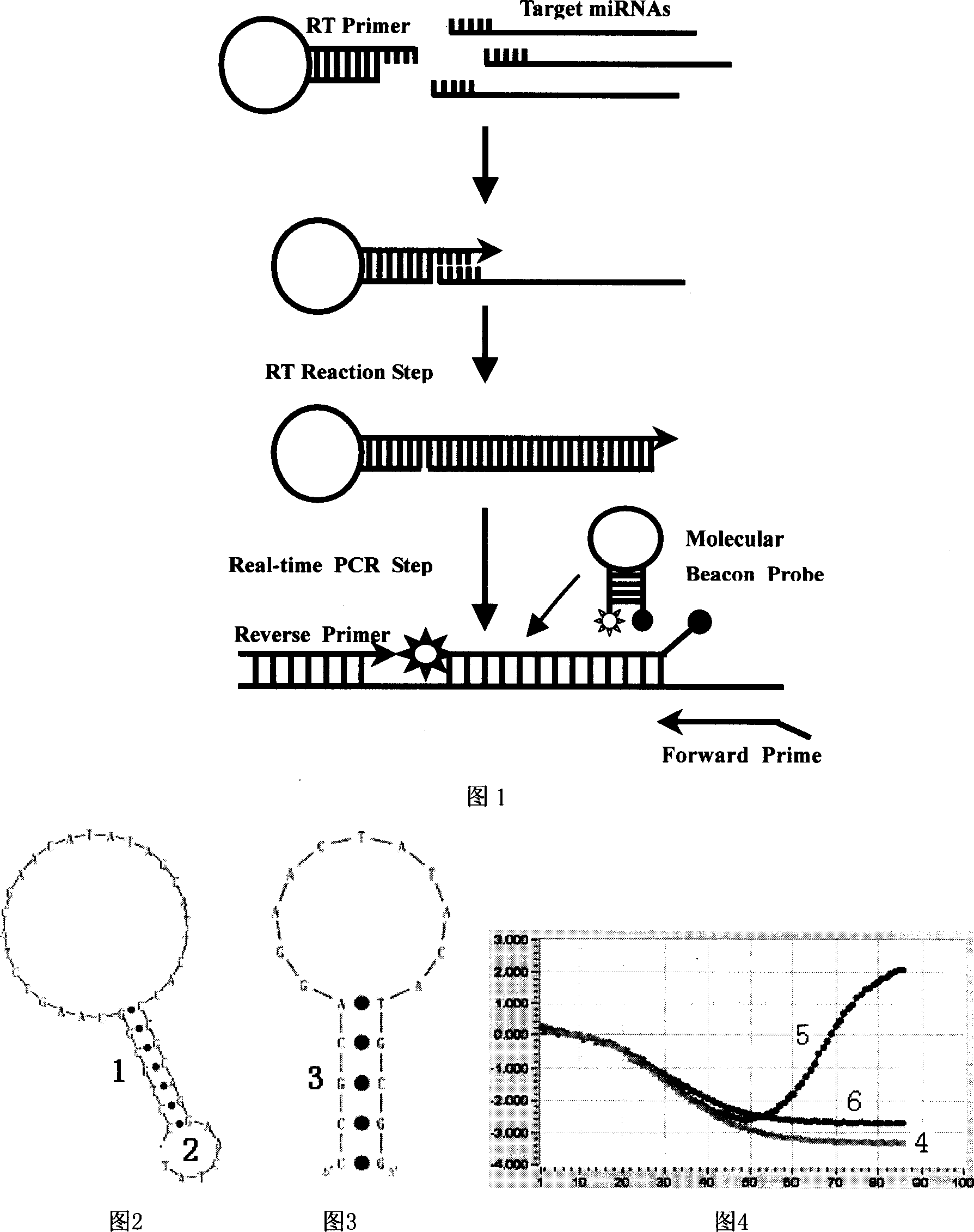
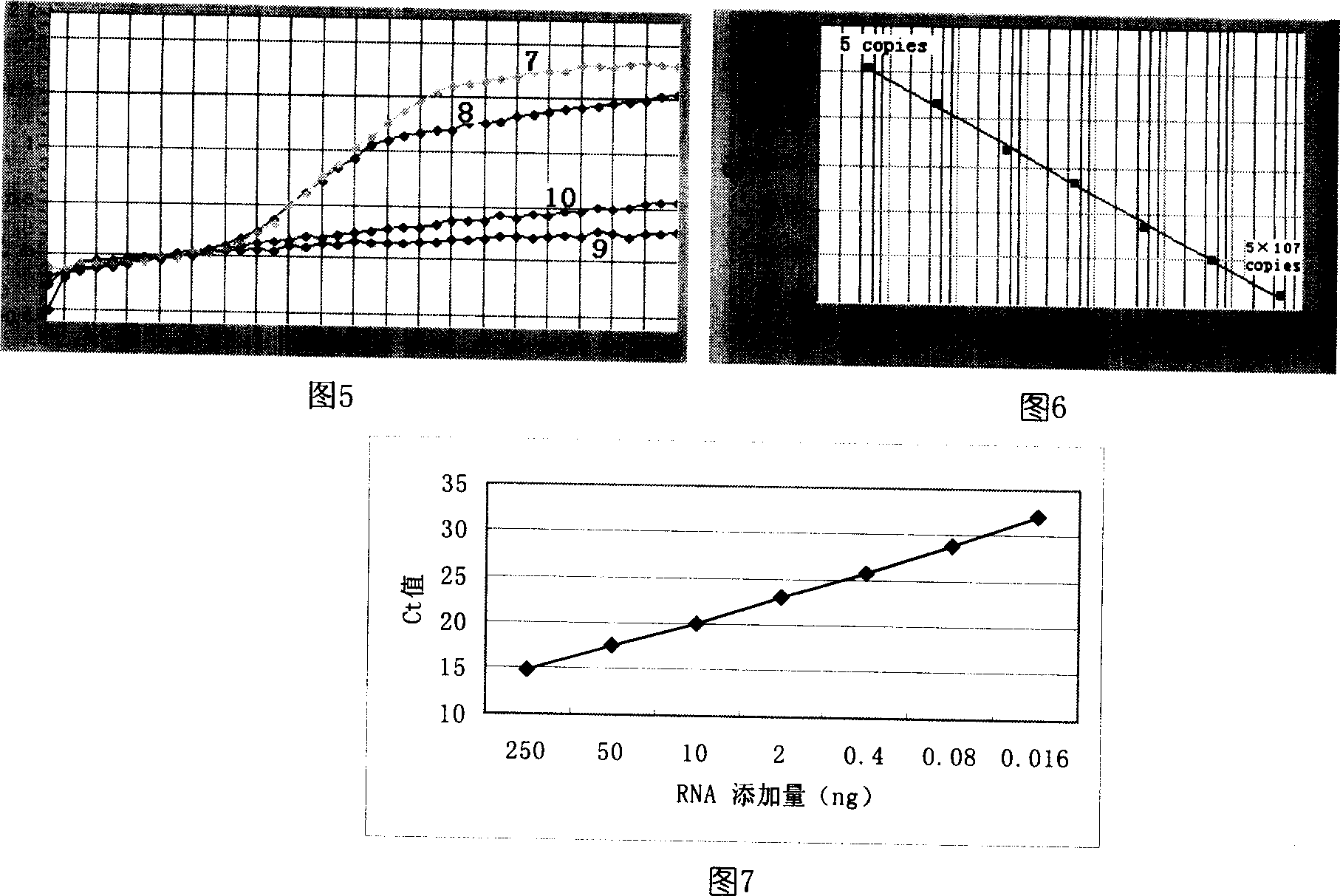
[0039] 1. Design of Let-7a and Let-7b primers and probes
[0040] Design the Let-7a RT primer as "5'-CCTGCGGCAAGTCTACGAACATATAGCATTCACCCGCAGGAACTAT-3'", as shown in Figure 2; where the base sequence "5'-CCTGCGG" at the 5' end is complementary to "CCGCAGG" in the "CACCCGCAGGAACTAT-3'" at the 3' end , so that the RT primer forms a stem-loop structure, and the 3' end "AACTAT-3" is complementary to the 3' end of Let-7a. The partial sequence "GGCAAGTCTACGAACAT" in the RT primer was used as one of the PCR primers, and the other PCR primer was designed according to the Let-7a sequence as "GCTTGAGGTAGTAGGTTG". The 3'-end "CCGCA" sequence of the RT primer is used as the 5'-end stem of the Let-7a molecular beacon probe, and the designed probe sequence is "FAM-5'-CCGCAGGAACTATACATGCGG-3'-DABCYL", as shown in Figure 3, where FAM is a fluorescent reporter group, and DABCYL is a fluorescent quencher group.
[0041]RT primer "5'-CCTGCGGCAAGTCTACGAACATATAGCATTCACCCGCAGGAACCAC-3'" and PCR pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com