Method and oligonucleotide for detecting FGFR3 gene G380R site mutation
An oligonucleotide and site mutation technology, which is applied in the field of highly sensitive detection of gene FGFR3G380R site mutation, can solve the problem of long distance between reporter fluorophore and quencher fluorophore, failure to recognize double-stranded DNA sequence, fluorescence quenching Incomplete extinction and other problems, to achieve the effect of long detection cycle, short detection cycle, simplified design and cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Oligonucleotides and kits for detecting mutations at the G380R site of the FGFR3 gene
[0046] The oligonucleotide used to detect the mutation of the G380R site of the FGFR3 gene in the sample, the oligonucleotide includes a pair of specific amplification products SEQ NO1 and SEQ NO2, and its base sequence is:
[0047] SEQ NO 1: GGGTTTTTCTCATCACTCTGCG
[0048] SEQ NO2: AGTGTGTATGCAGGCATCCT,
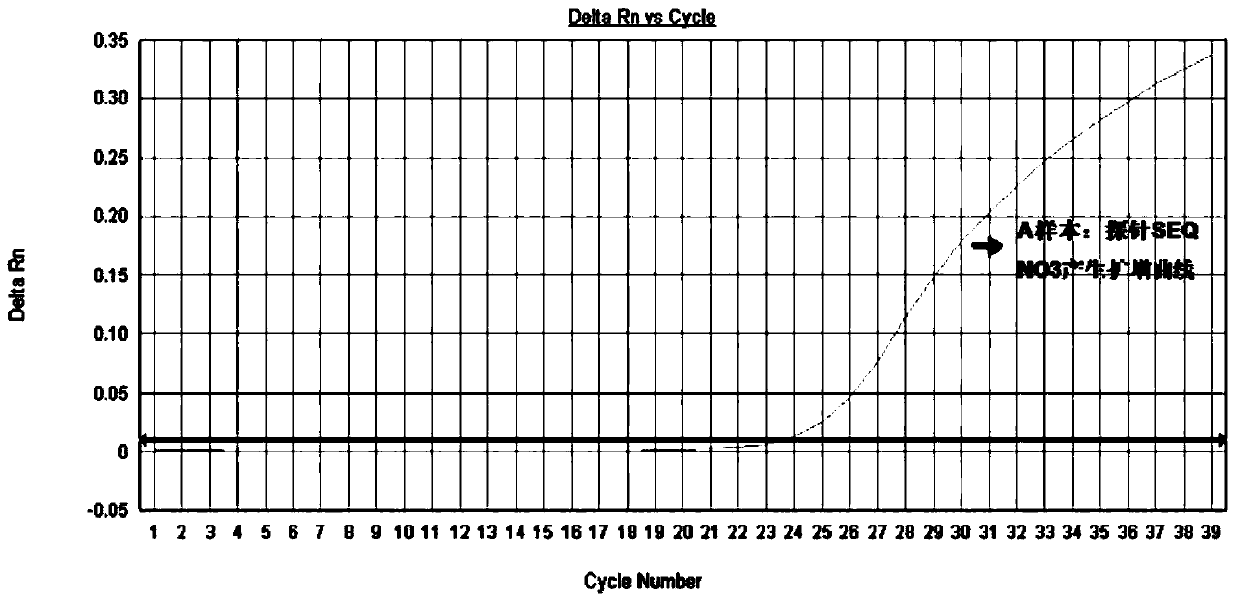
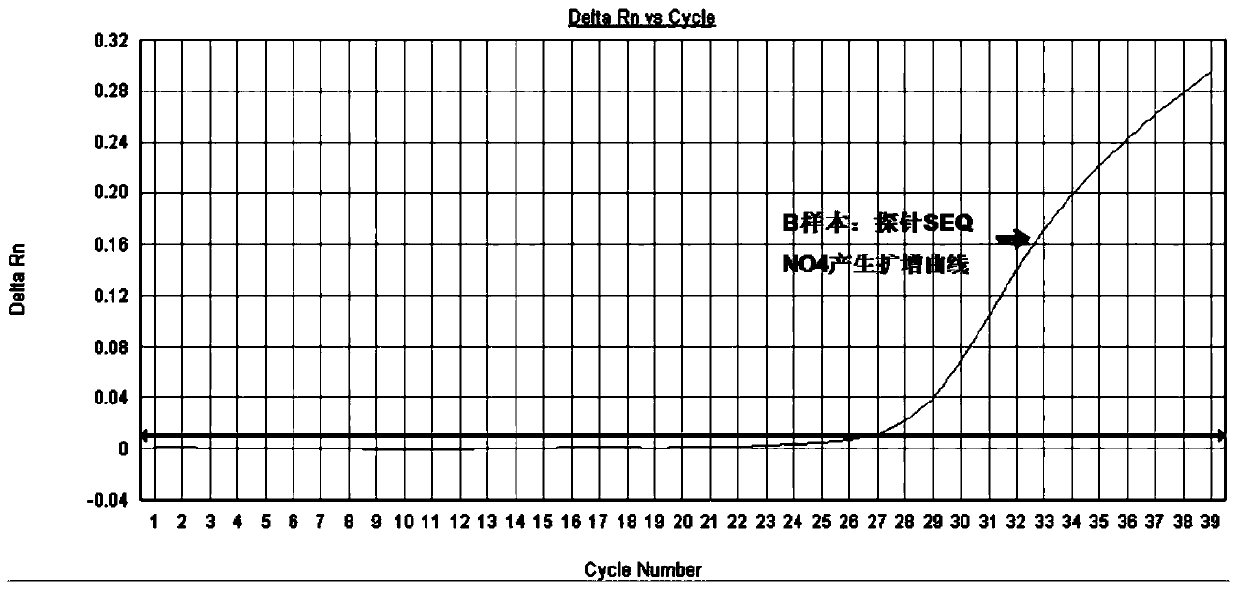
[0049] And a pair of specific detection probes SEQ NO3 and SEQ NO4, its base sequence is:
[0050] SEQ NO 3: FAM-AAGAAGCCCACCCCGTAGCT-MGB
[0051] SEQ NO4: HEX-AAGAAGCCCACCCTGTAGCT-MGB
[0052] Among them, MGB is marked at the 3' end of SEQ NO3 and SEQ NO4 to enhance detection specificity.
[0053] A kit for detecting mutations at the G380R site of the FGFR3 gene in a sample, the kit including sample DNA extraction reagents, erythrocyte lysate, absolute ethanol, qPCR amplification reaction solution, positive control substance, negative control substance and blank cont...
Embodiment 2
[0063] Embodiment 2: detection process
[0064] (1) Blood DNA extraction: Take 500ul whole blood, put it into a 1.5ml centrifuge tube, add 1ml red blood cell lysate. Turn it upside down to make it completely mixed, rotate and pulsate for 15 seconds, and then put it into a centrifuge for centrifugation at 5000rpm for 10min. Pour off the upper layer, and it can be seen that there is a bloody precipitate at the bottom of the centrifuge tube. Add 500ul red blood cell lysate, repeat this lysis step once. Centrifuge at 5000rpm for 5min, and finally suck up all the upper layer with a pipette and discard it, so that the blood-colored precipitate at the bottom of the centrifuge tube no longer has lysate. Mix well with a metal bath at 100°C for 10 minutes, 12000 rpm for 5 minutes, take the supernatant, and obtain the sample DNA solution. When not used immediately, the sample DNA solution was stored at -20°C until use.
[0065] (2) Real-time fluorescent PCR amplification:
[0066] T...
Embodiment 3
[0070] Embodiment 3: Sample detection verification
[0071] 7 clinical samples were selected, numbered 1-7, among which samples 1-5 were clinically diagnosed as patients with ACH, sample 6 was from the wife of patient 5, the wife of patient 5 did not suffer from ACH, and had been pregnant for 20 weeks, and sample 7 was number 5 Amniotic fluid from the patient's wife. The implementation steps are as follows:
[0072] (1) Samples 1-6 are peripheral blood. DNA was extracted according to the blood DNA extraction method described in Example 2. Sample 7 was extracted from chorion exfoliated cells in amniotic fluid using the QIAamp DNA extraction kit from QIAGEN.
[0073] (2) According to the real-time fluorescent PCR amplification method described in Example 2, real-time fluorescent PCR detection was performed on the DNA of samples 1-7, and Sanger sequencing analysis was performed on the 10th exon of the FGFR3 gene of these 7 samples. The results of real-time fluorescent PCR (qPCR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com