Method for identifying significantly different expressed genes
A technology of differentially expressed genes and collections, which is applied in biochemical equipment and methods, microbial measurement/inspection, special data processing applications, etc., can solve the problems of low accuracy in identifying significantly expressed gene collections and excessive detection times, and achieve Excellent recognition effect, value-enhancing, and high-accuracy effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0026] Below in conjunction with accompanying drawing, the invention is further explained.
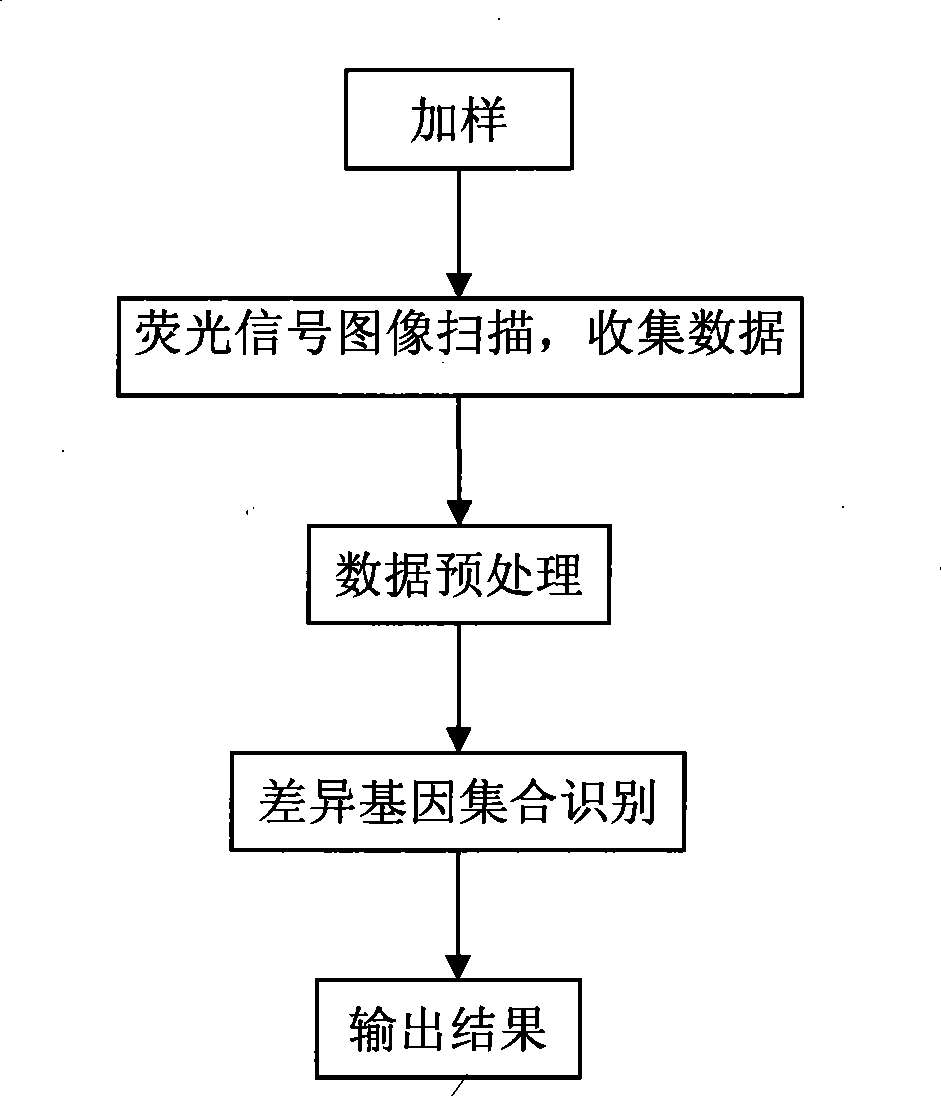
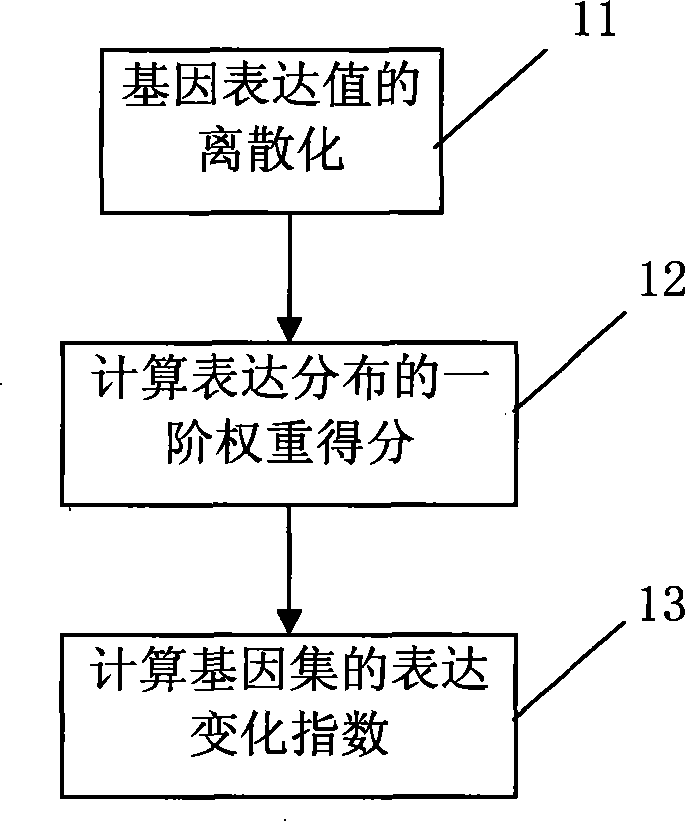
[0027] Such as figure 1 As shown, after the sample is fluorescently labeled, the sample is added to the high-throughput biochip gene expression detection and identification device, and hybridized with the probe on the gene chip. After the hybridization is completed, it is cleaned to remove impurities, and the photoelectric part detects and records the gene expression. Intensity, preprocess the data to generate gene expression microarray data. Such as figure 2 , which are then processed by the identification mechanism to identify sets of genes with significant expression changes.
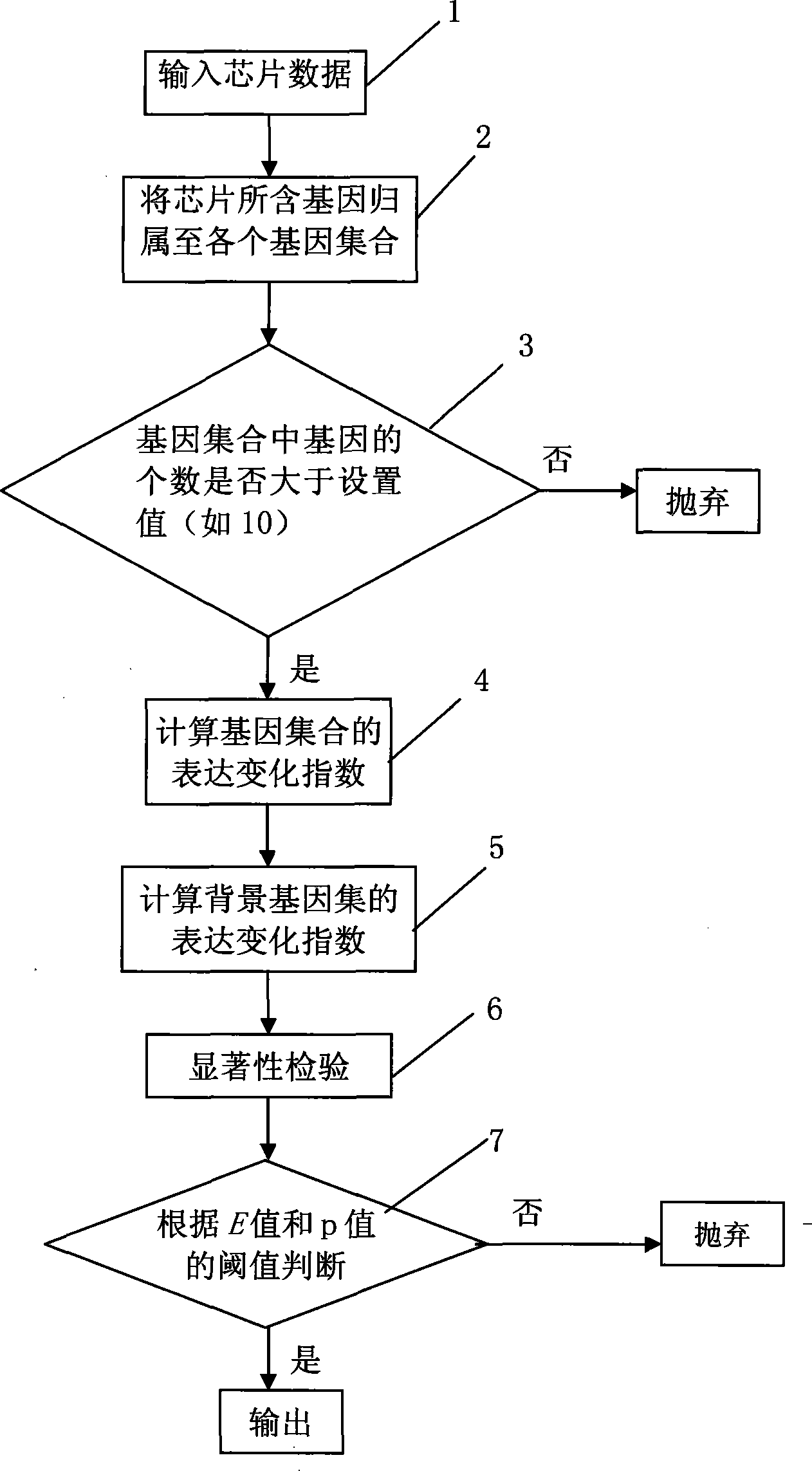
[0028] The method of the present invention is as figure 2 shown. Step 1 is the initial action, including user settings and input data; Step 2 assigns the genes contained in the chip to each gene set; Step 3 judges whether the number of genes in each gene set is greater than the number defined in the set...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com