Detection and quantification of biomolecules using mass spectrometry
A technology of biomolecules and mass spectrometry, applied in biochemical equipment and methods, microbial measurement/testing, fermentation, etc., can solve time-consuming problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0144] The examples provided below illustrate but do not limit the invention.
example 1
[0145] Example 1: Detection of exon 10 of the RhD gene
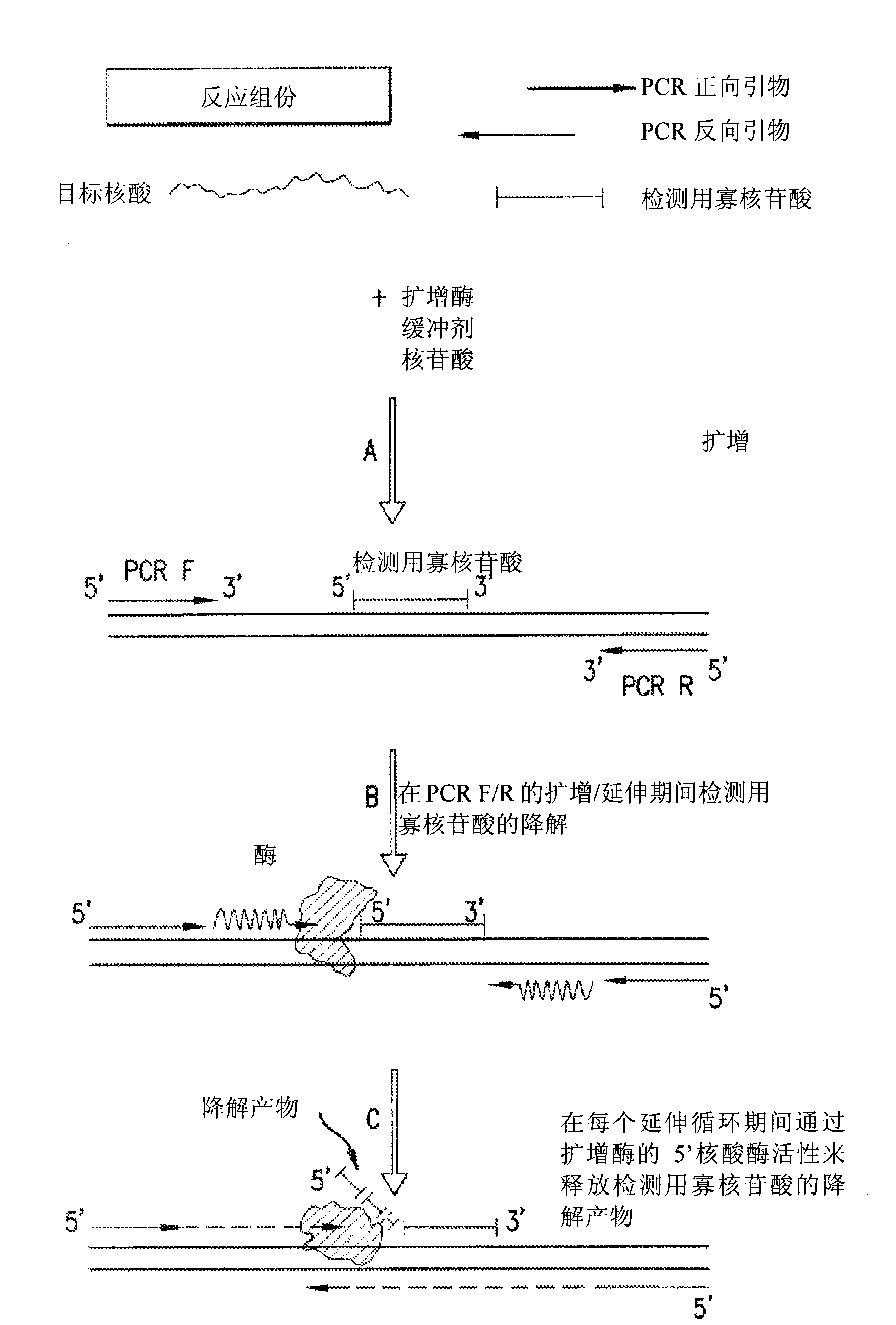
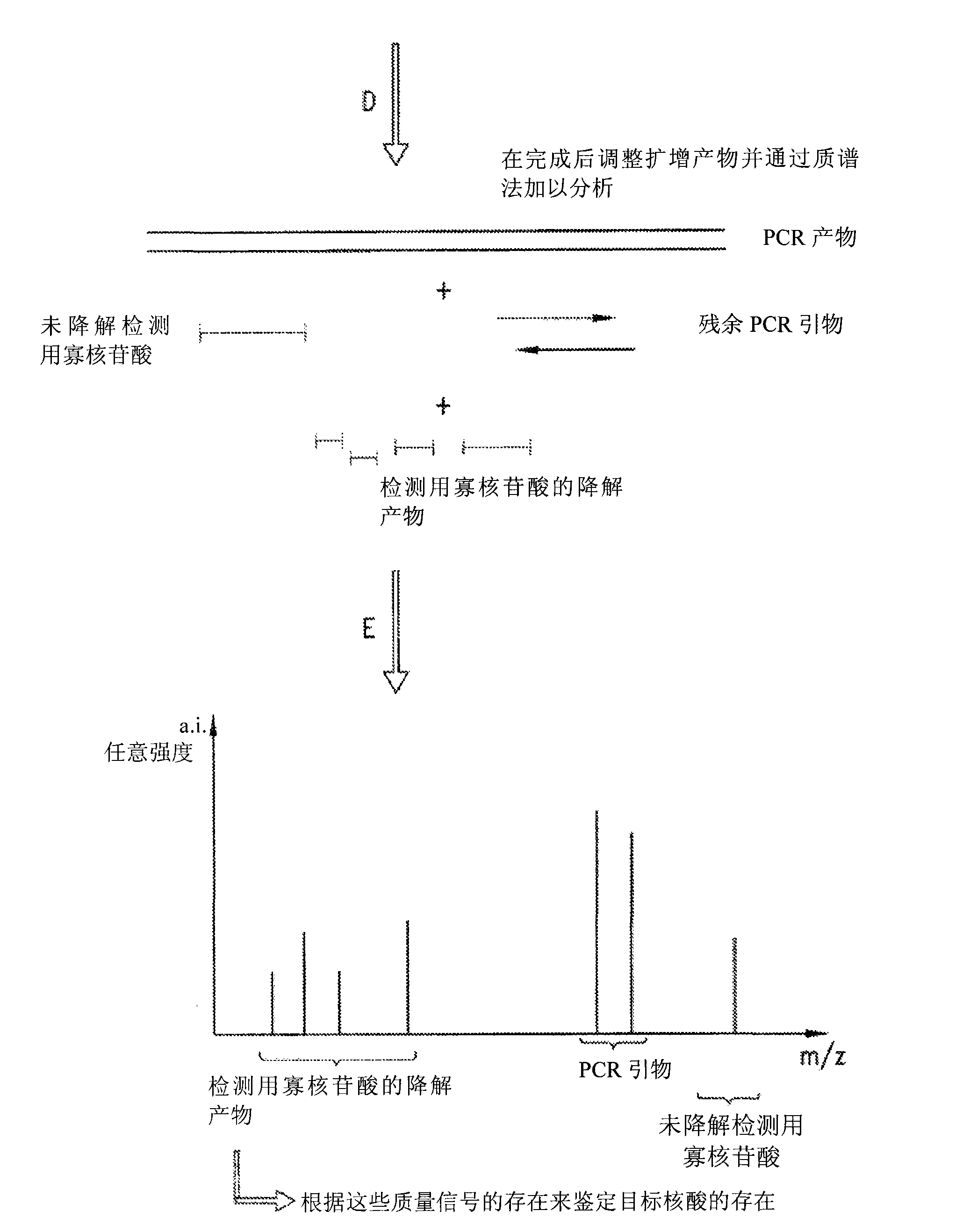
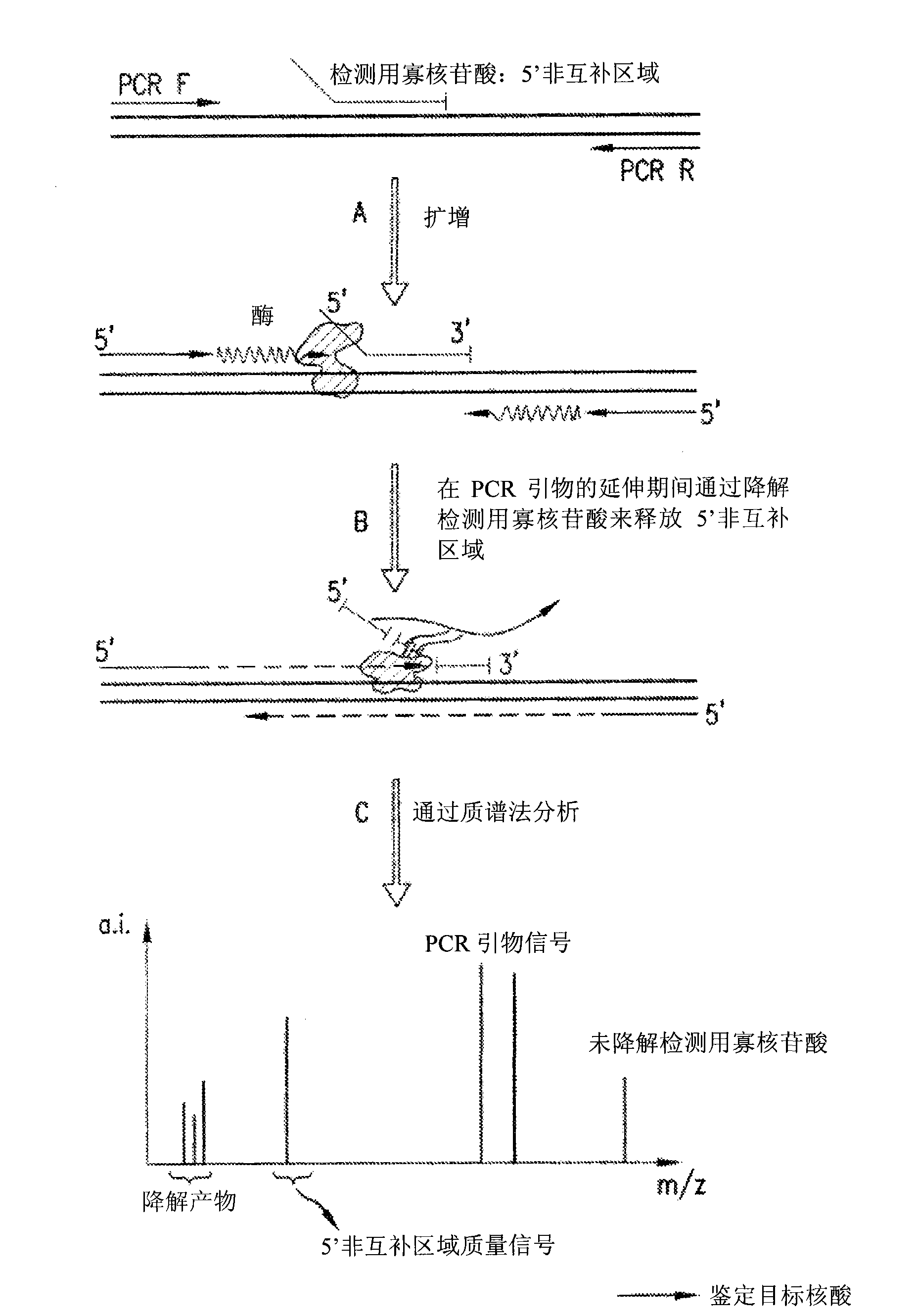
[0146] Detection assays were performed to detect the exon 10 region of the rhesus monkey D gene. The design of PCR primers and detection oligonucleotides was carried out according to the specific instructions section. In a particular assay, the detection oligonucleotide carries a non-complementary 5' overhang consisting of 6 adenines. Due to the presence of the target sequence (exon 10 of RhD) in the sample, the PCR primers and detection oligonucleotides hybridize to the target. During amplification, the detection oligonucleotide is degraded by the 5' nuclease activity of the DNA polymerase extending upstream from the PCR primer. During degradation, mass-distinguishable products (MDPs) including the 5' polyA tag are released and unambiguously identified by mass spectrometry (see Figure 8 ). Detection of these mass signals confirms the presence of the target nucleic acid.
[0147] Primers and detection oligon...
example 2
[0164] Example 2: Detection of RhD gene exon 5
[0165] Detection assays were performed to detect the exon 5 region of the rhesus monkey D gene. The design of PCR primers and detection oligonucleotides was carried out according to the specific instructions section. In a particular assay, the detection oligonucleotide carries a non-complementary 5' overhang consisting of 8 adenines. Due to the presence of the target sequence (exon 5 of RhD) in the sample, the PCR primers and detection oligonucleotides hybridize to the target. During amplification, the detection oligonucleotide is degraded by the 5' nuclease activity of the DNA polymerase extending upstream from the PCR primer. During degradation, mass-distinguishable products (MDPs) including the 5' polyA tag are released and unambiguously identified by mass spectrometry (see Figure 9 ). Detection of these mass signals confirms the presence of the target nucleic acid.
[0166] Primers and detection oligonucleotide ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com