microRNA target position point prediction method based on support vector machine
A technology of support vector machines and target sites, which is used in the determination/inspection of microorganisms, biochemical equipment and methods, special data processing applications, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
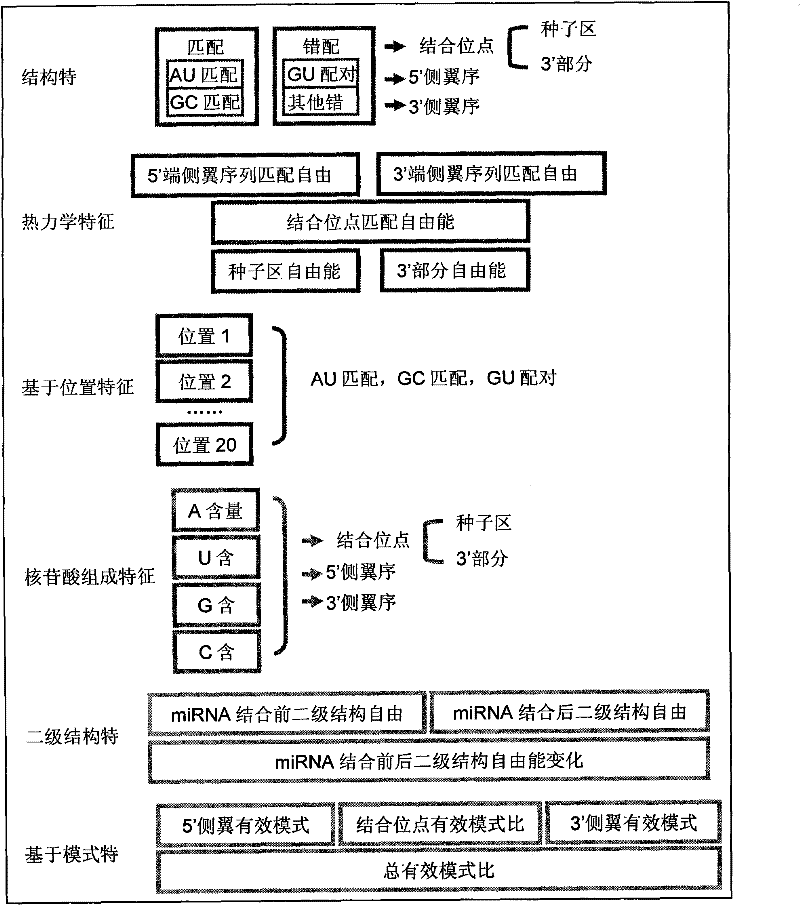
[0061] The method for the microRNA target site prediction based on support vector machine, comprises the steps:
[0062] 1. Establishment of training data set
[0063] Since the training set data is very important for machine learning methods, selecting appropriate positive and negative sample sets is one of the key points and difficulties of this study. Using the miRecords database, miRecordsversion 1[9] has a total of 1979 records, including 121 records for fruit flies and 1311 records for humans; only the data of these two animals are taken as data sets. After removing duplicate records and records with incomplete information (mainly because the position of the binding site is not given), a total of 278 miRNA-target site interaction pairs were obtained, of which 83 were from Drosophila and 195 were from human , these samples are used as positive samples.
[0064] Negative samples are often more important than positive samples for the specificity of a classifier. Previous...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com