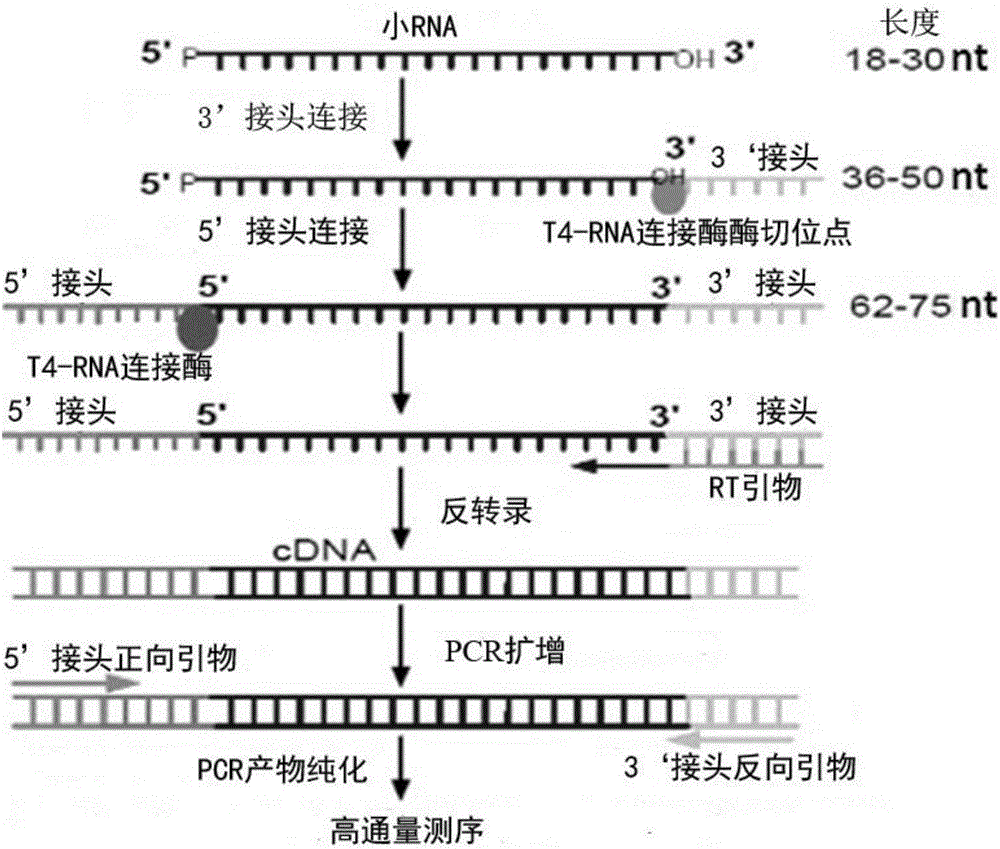
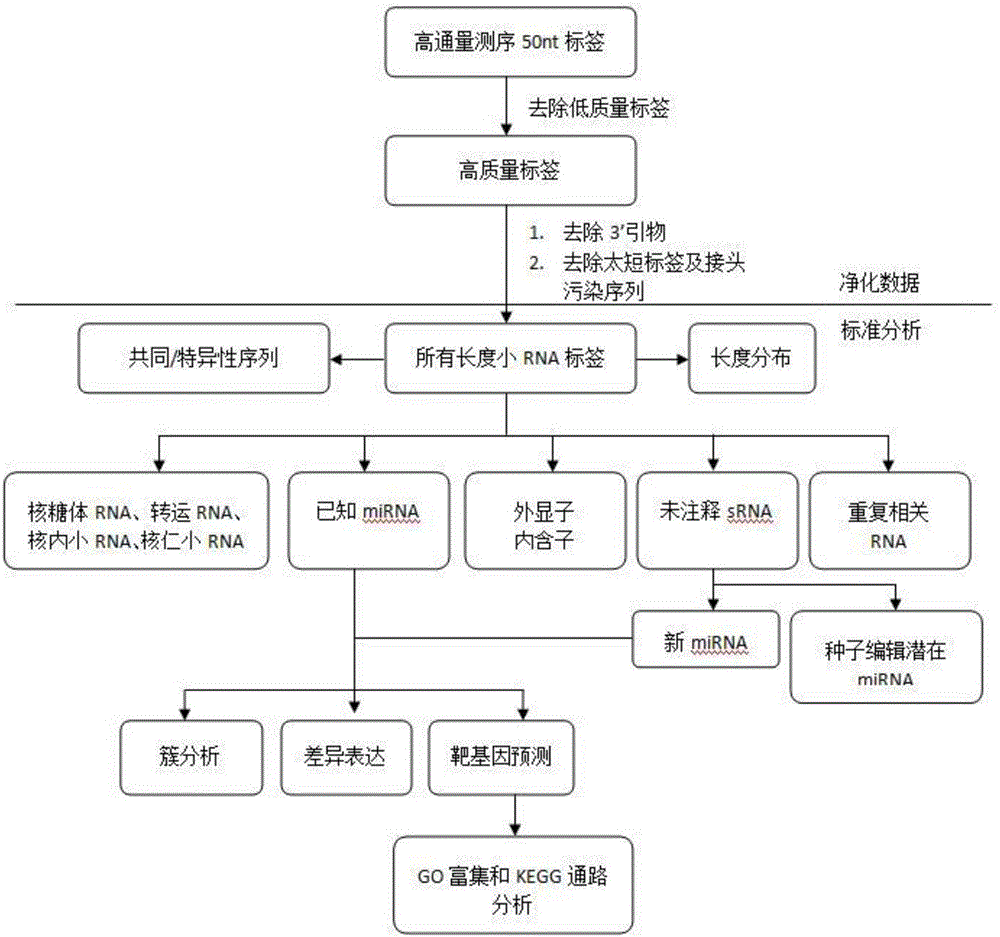
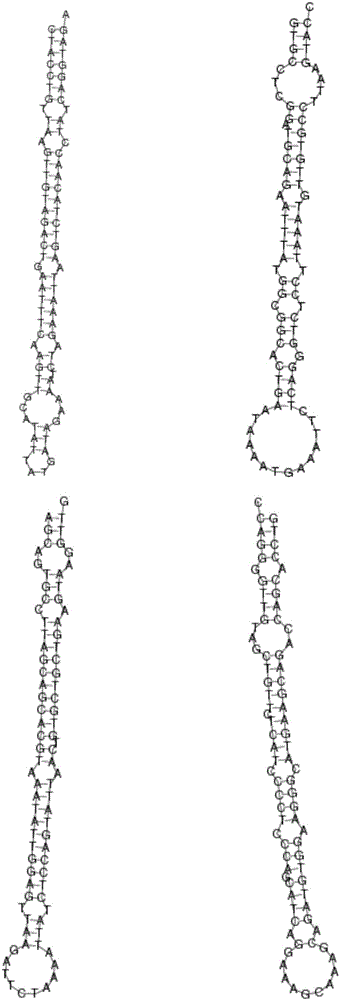
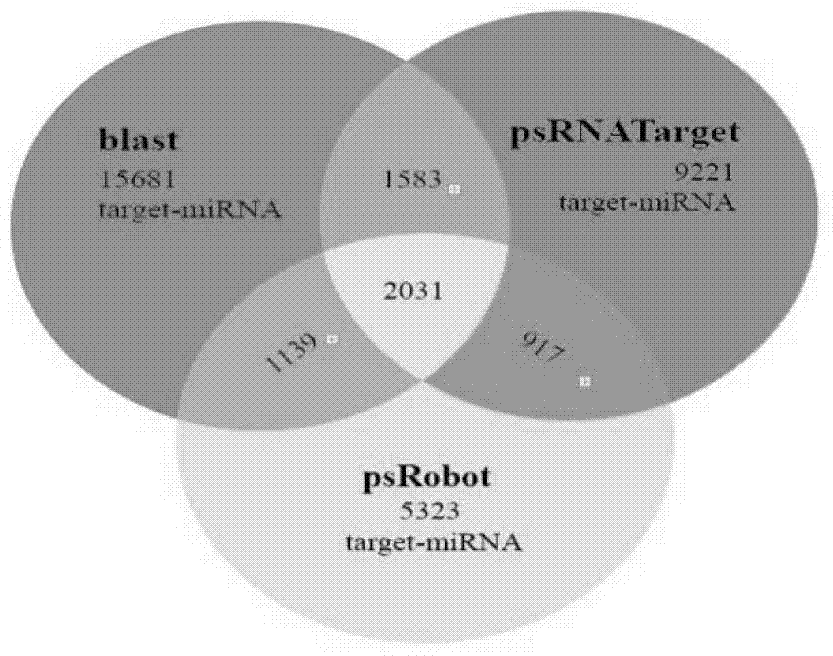
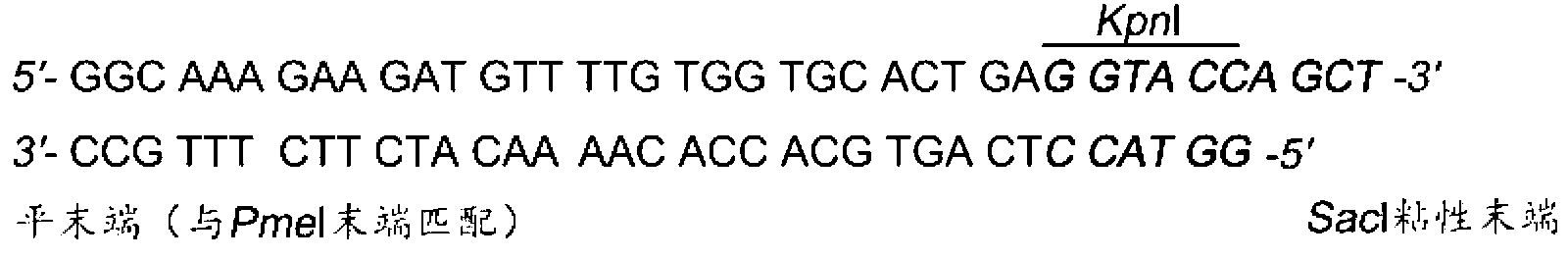Patents
Literature
72 results about "Mirna target" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Pharmaceutical Composition
InactiveUS20100004320A1Reduce depressionEffectively repressedAntibacterial agentsOrganic active ingredientsNucleotideNucleobase
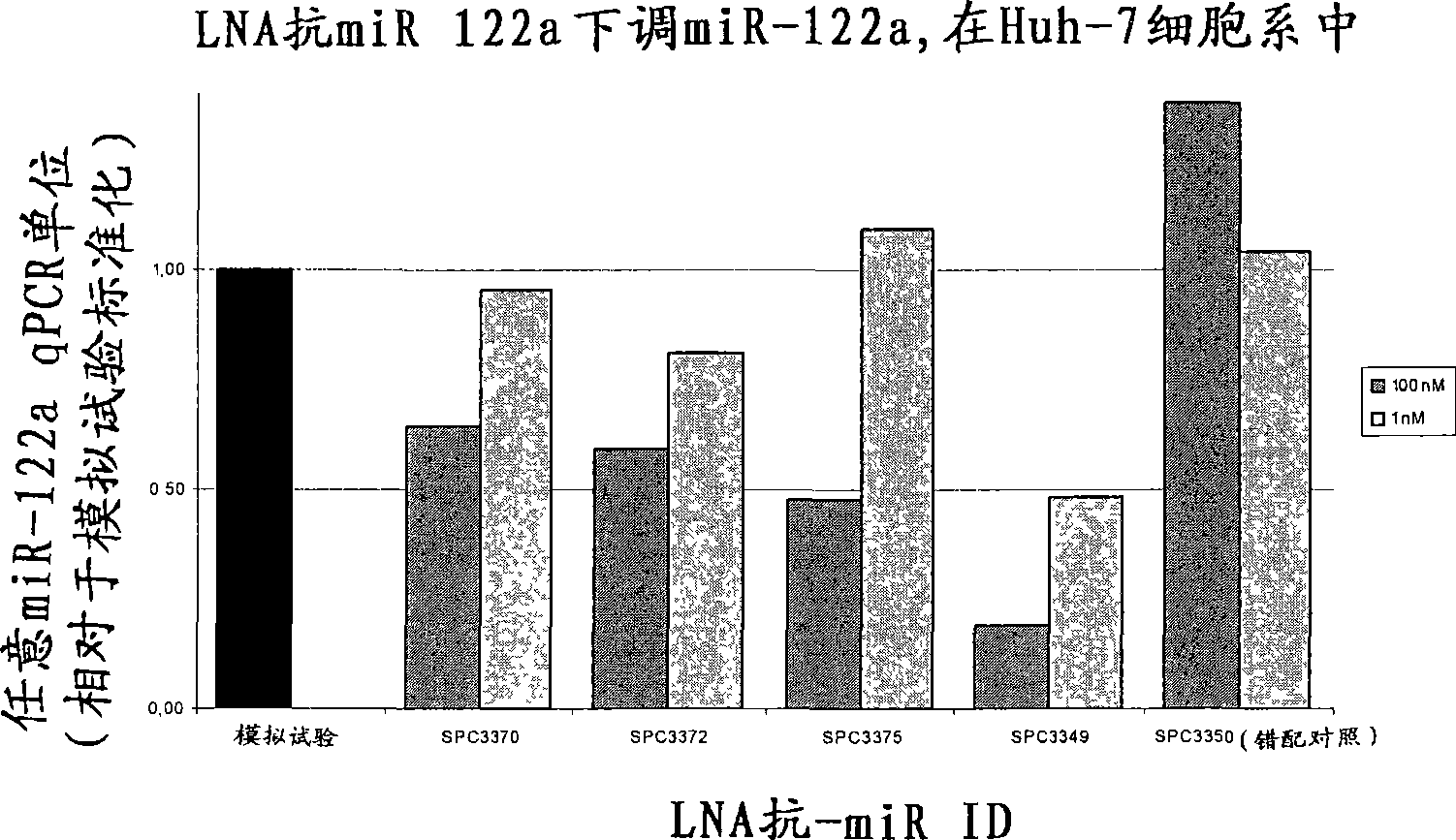
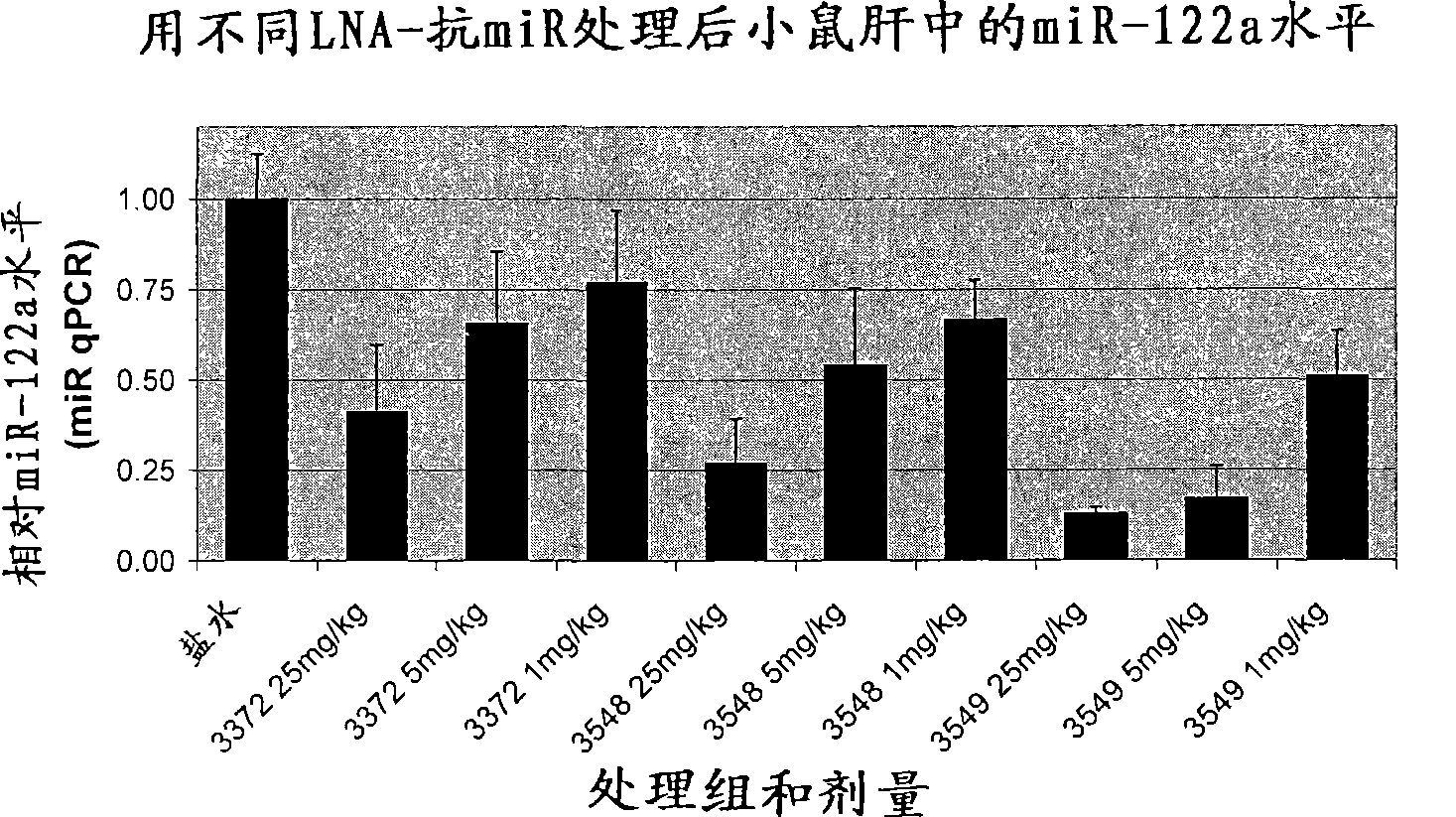
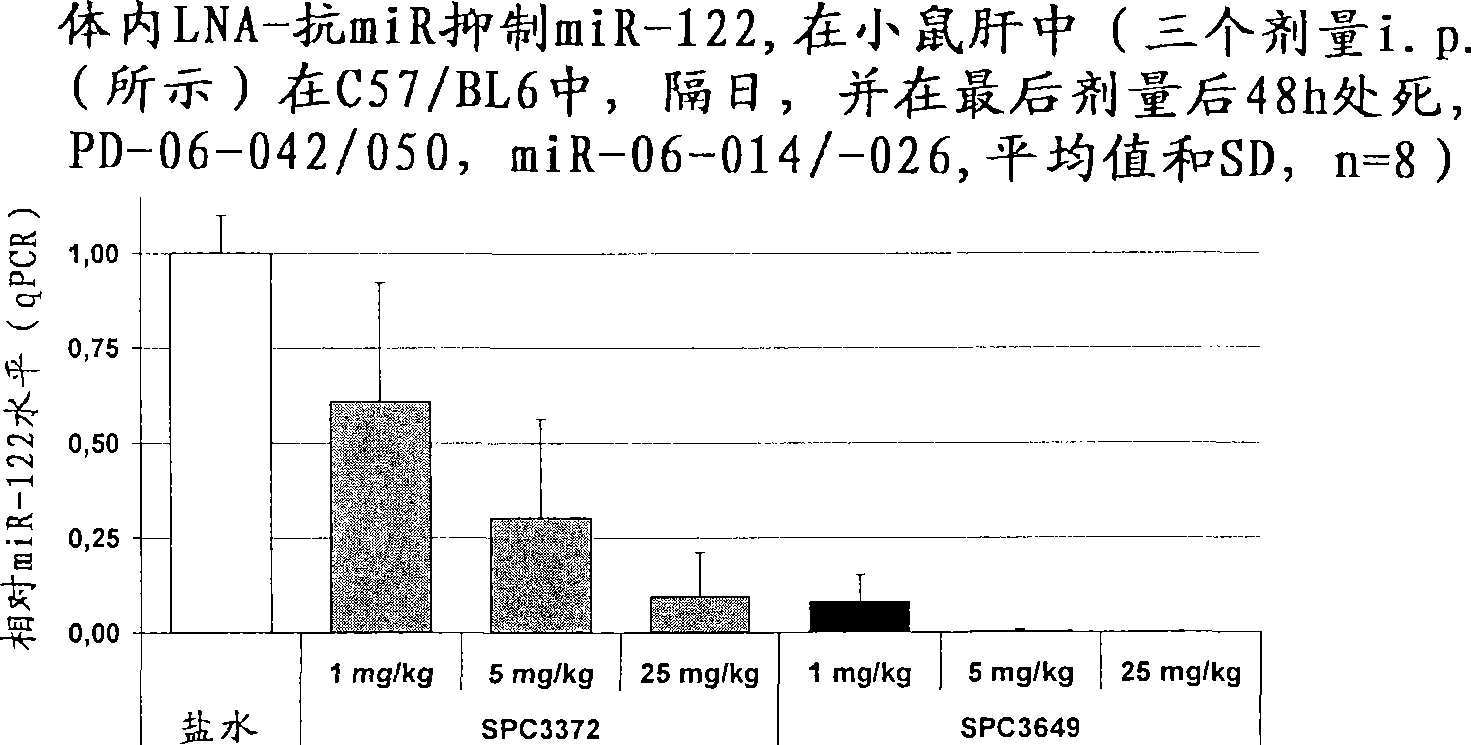
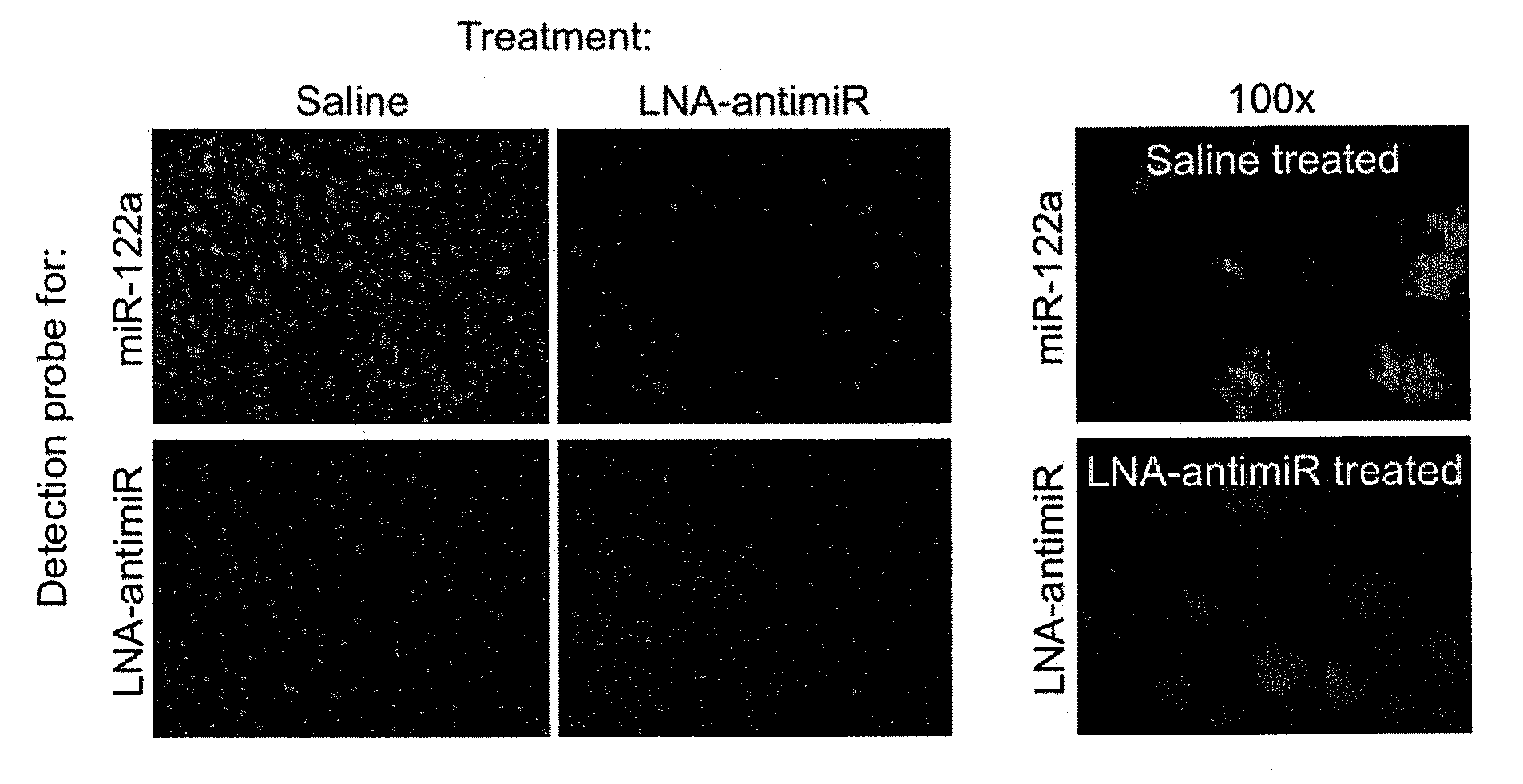
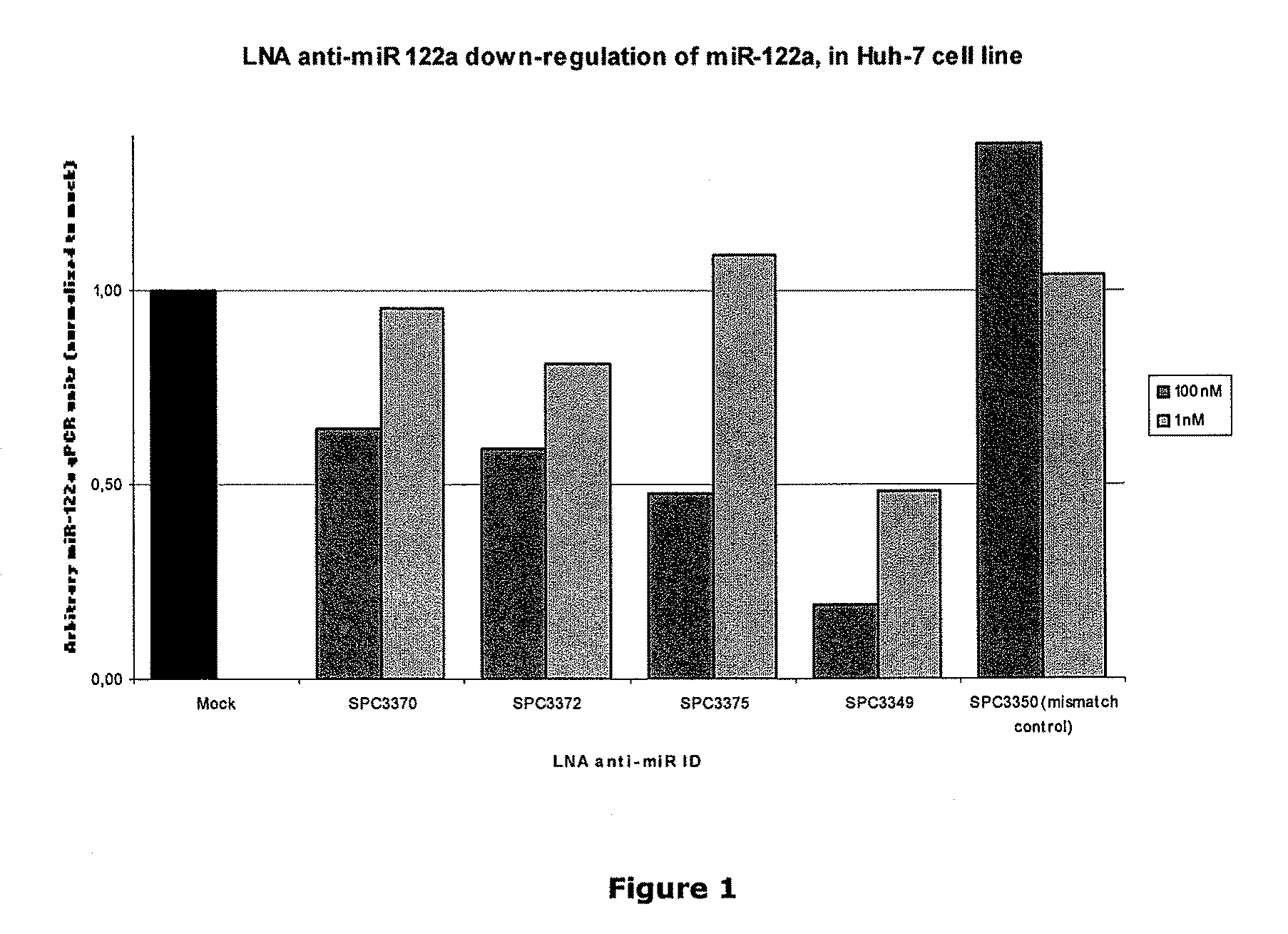
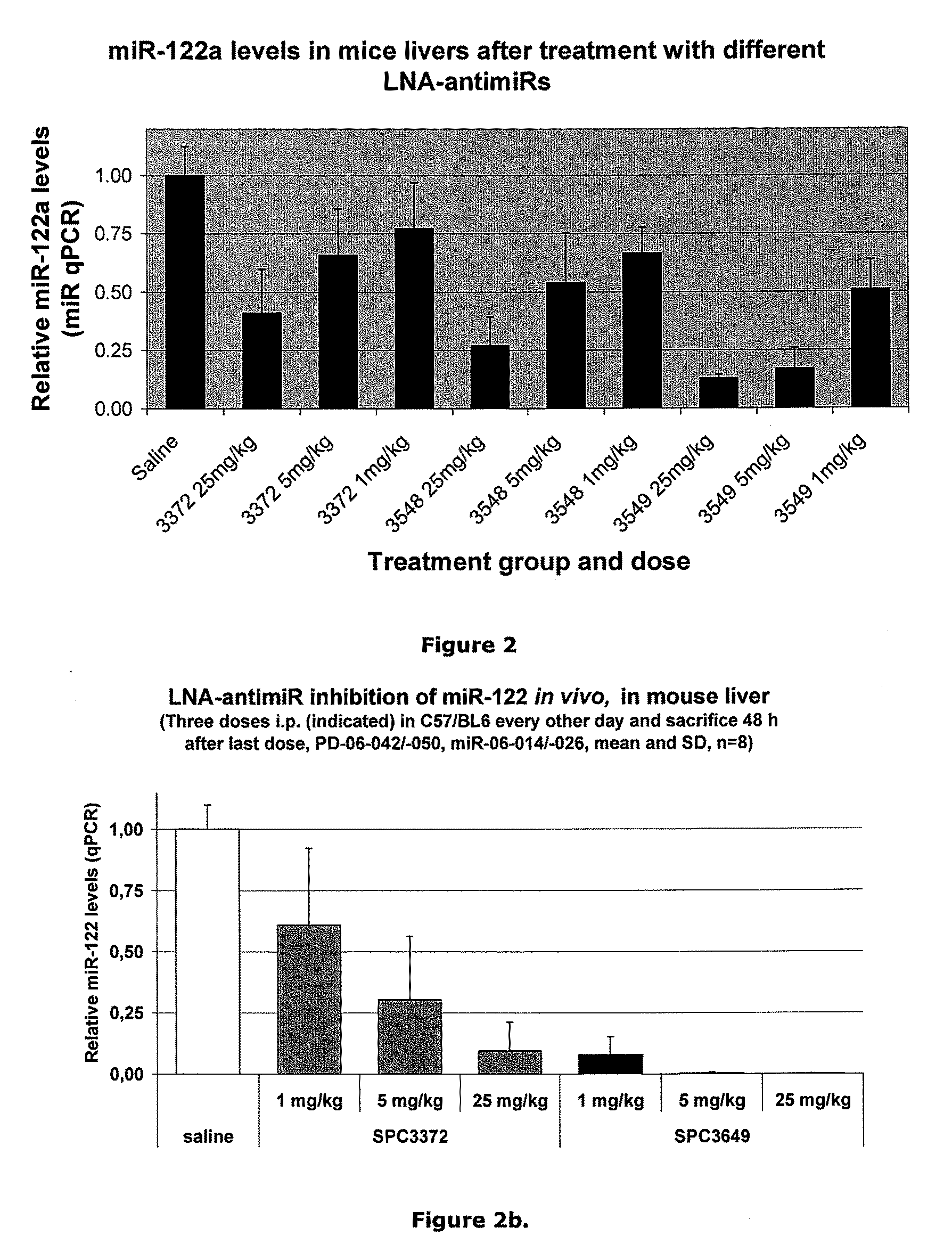
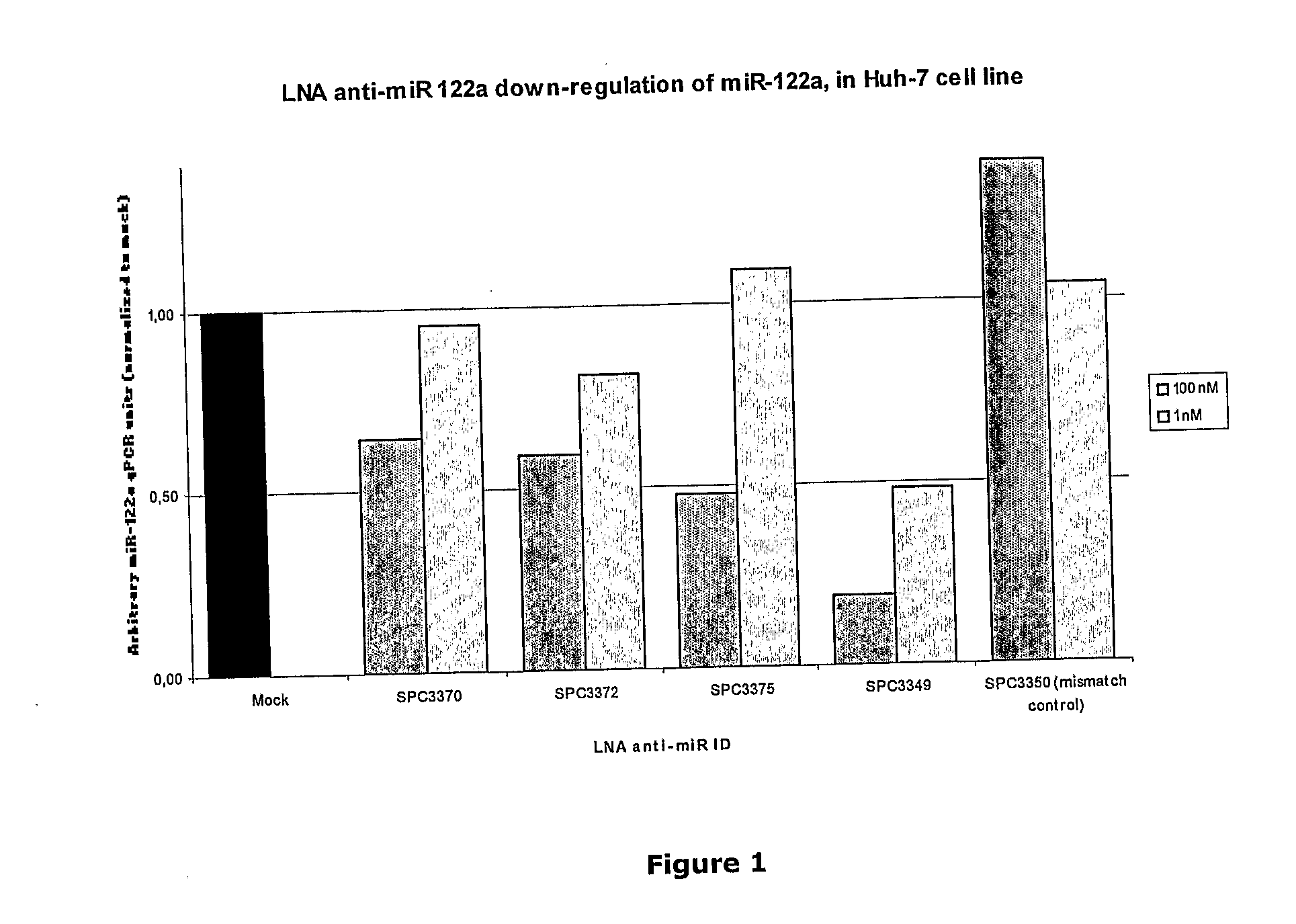
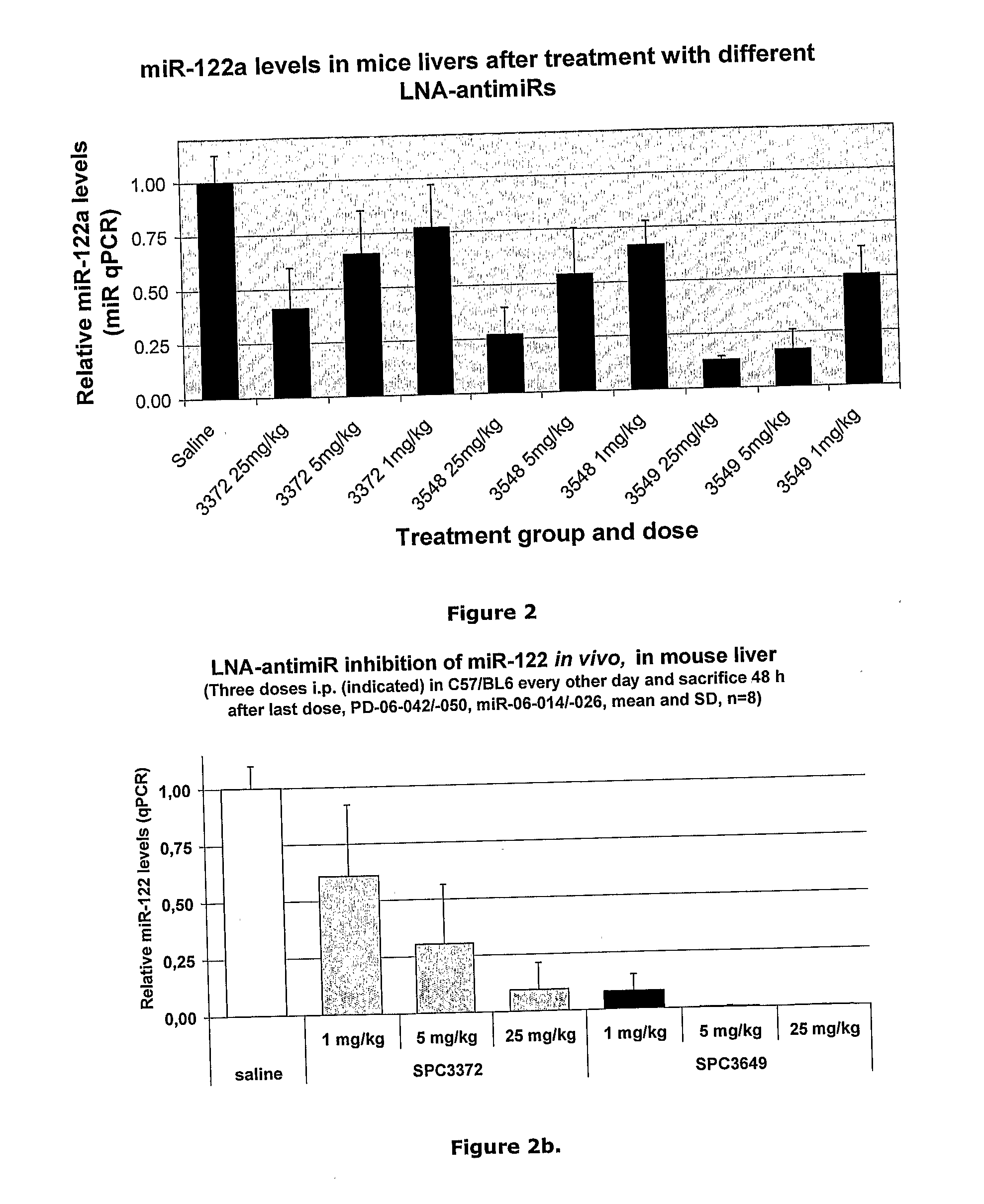
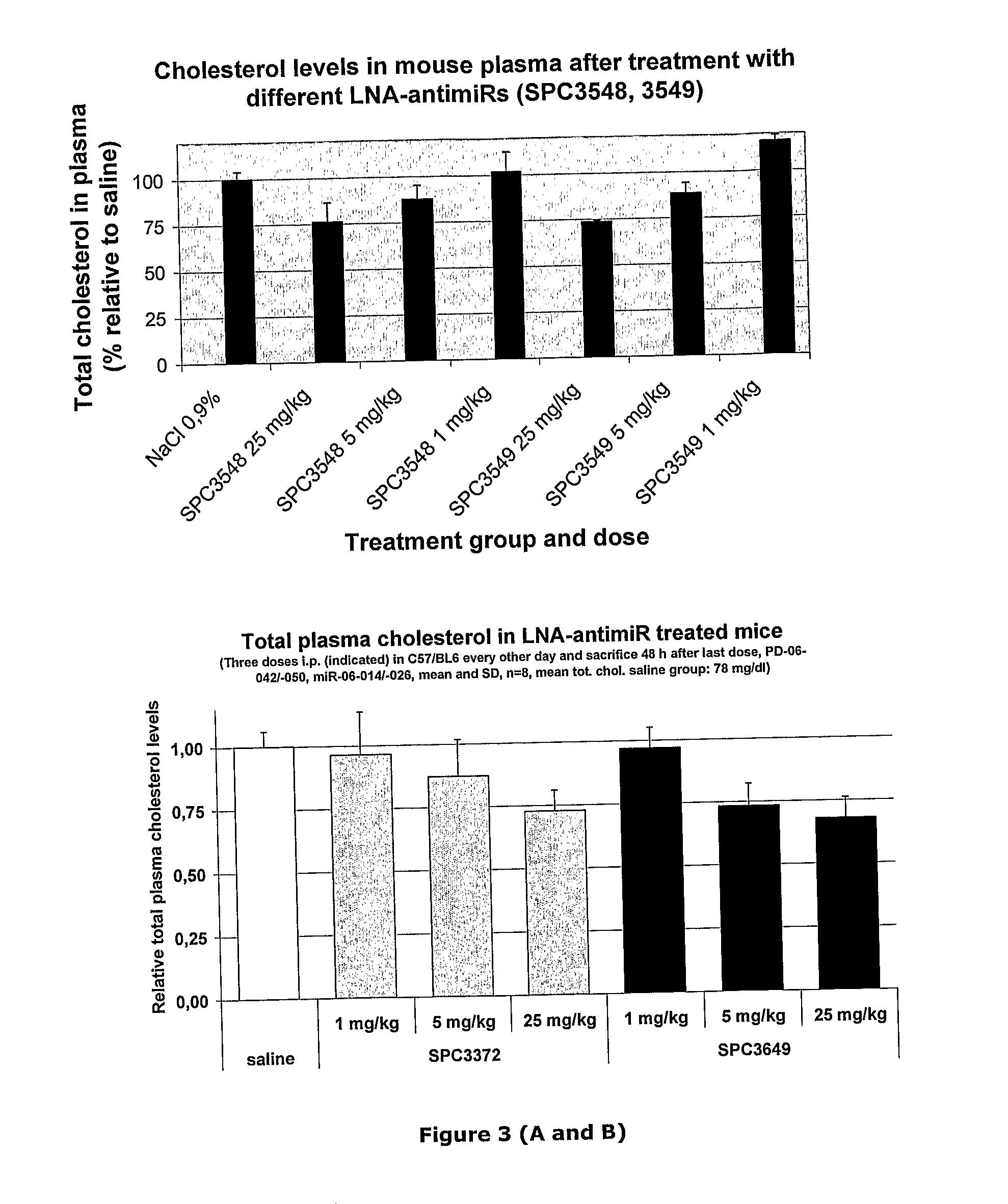
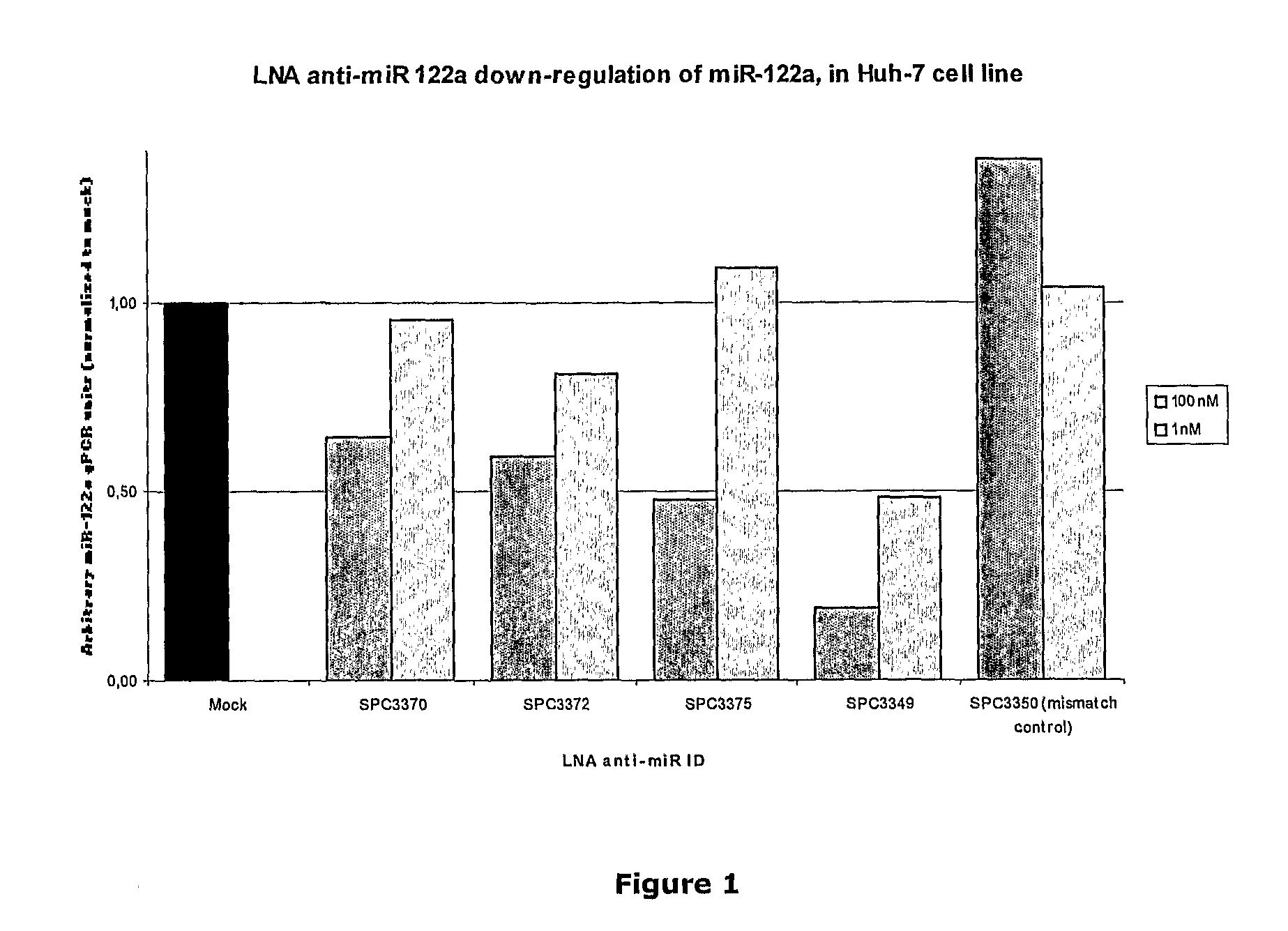
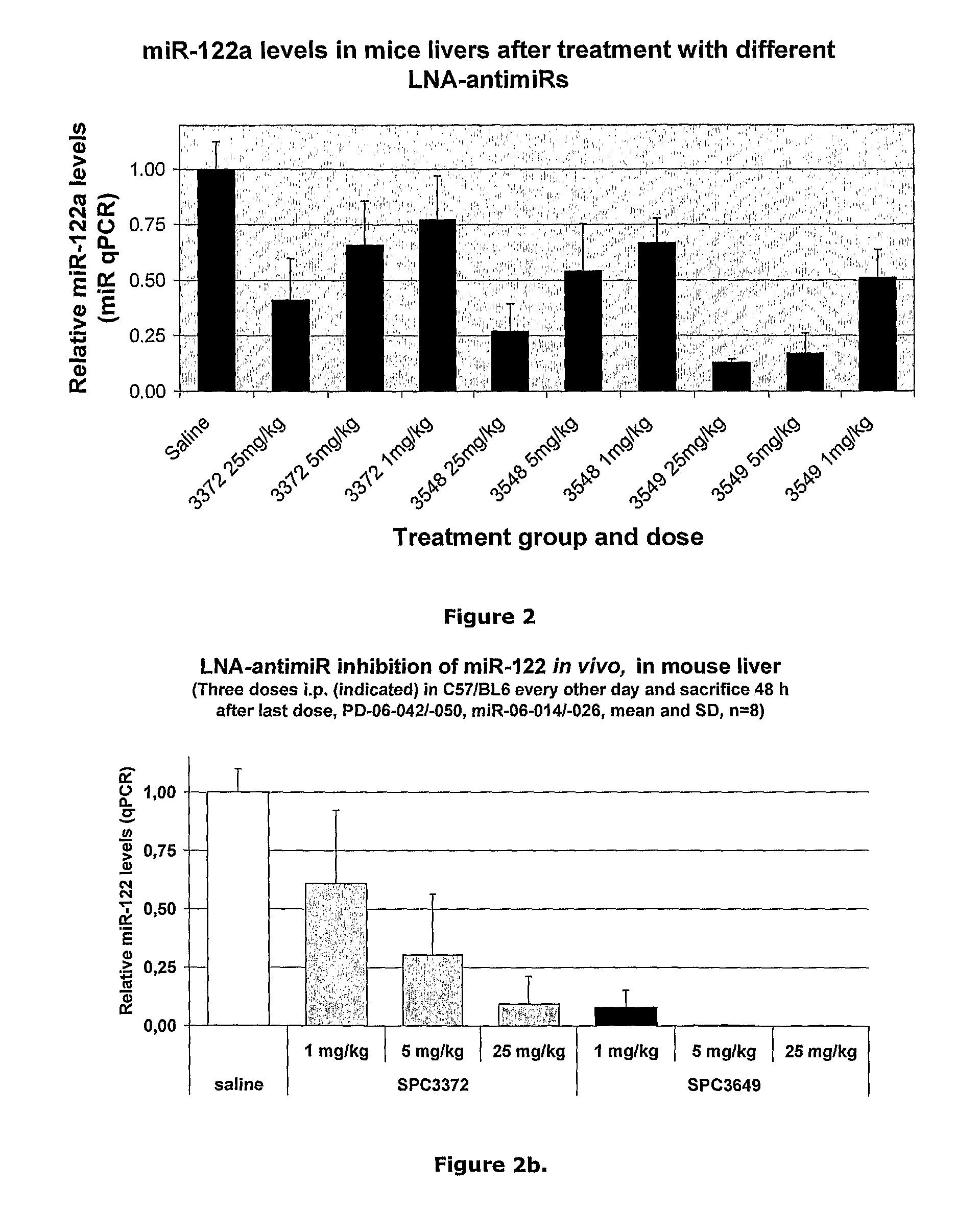
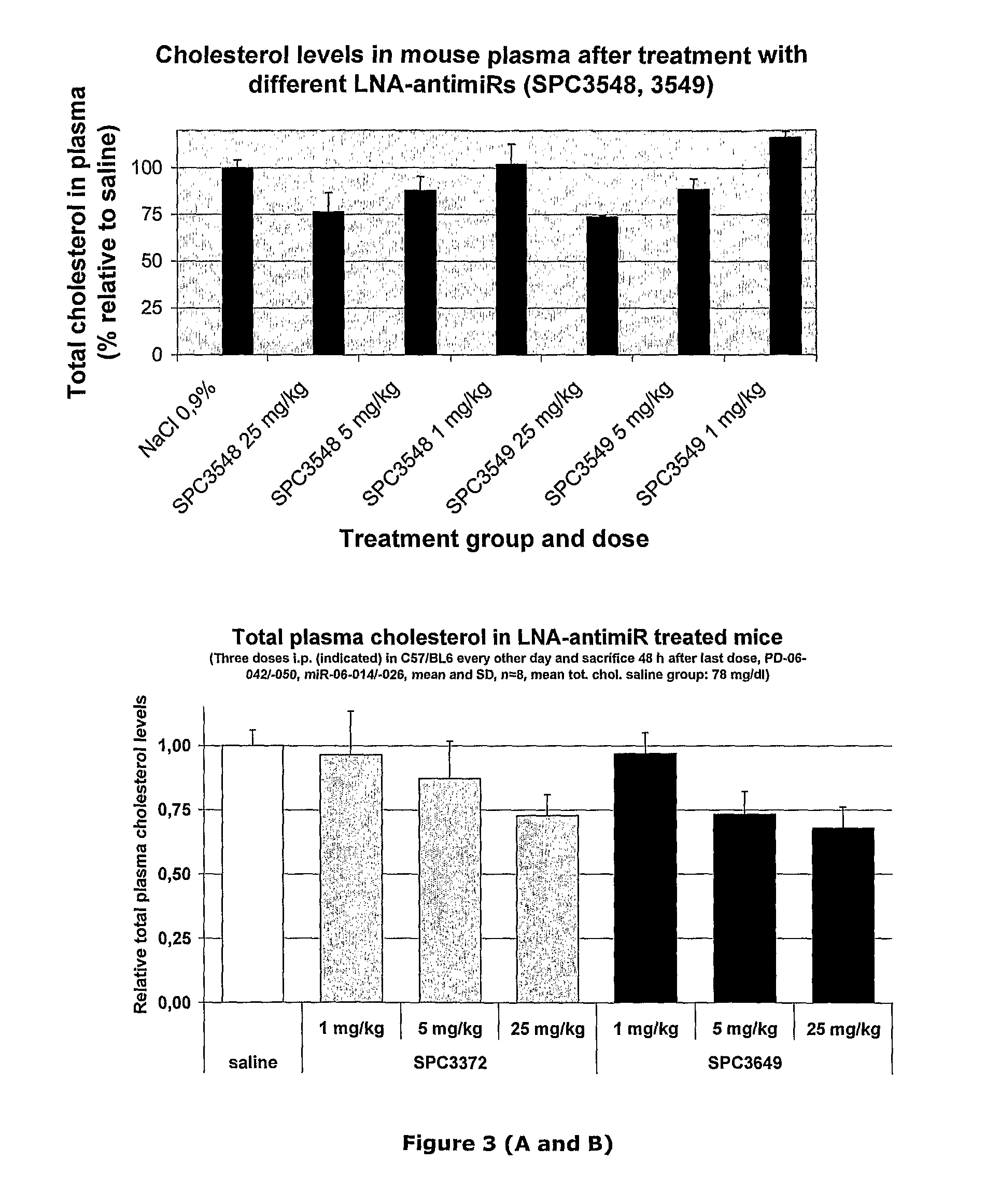
The invention provides pharmaceutical compositions comprising short single stranded oligonucleotides, of length of between 8 and 17 nucleobases which are complementary to human microRNAs. The short oligonucleotides are particularly effective at alleviating miRNA repression in vivo. It is found that the incorporation of high affinity nucleotide analogues into the oligonucleotides results in highly effective anti-microRNA molecules which appear to function via the formation of almost irreversible duplexes with the miRNA target, rather than RNA cleavage based mechanisms, such as mechanisms associated with RNaseH or RISC.
Owner:SANTARIS PHARMA AS
Methods and compositions for gene silencing
InactiveUS20070130653A1Sugar derivativesOther foreign material introduction processesDevelopmental stageHeterologous
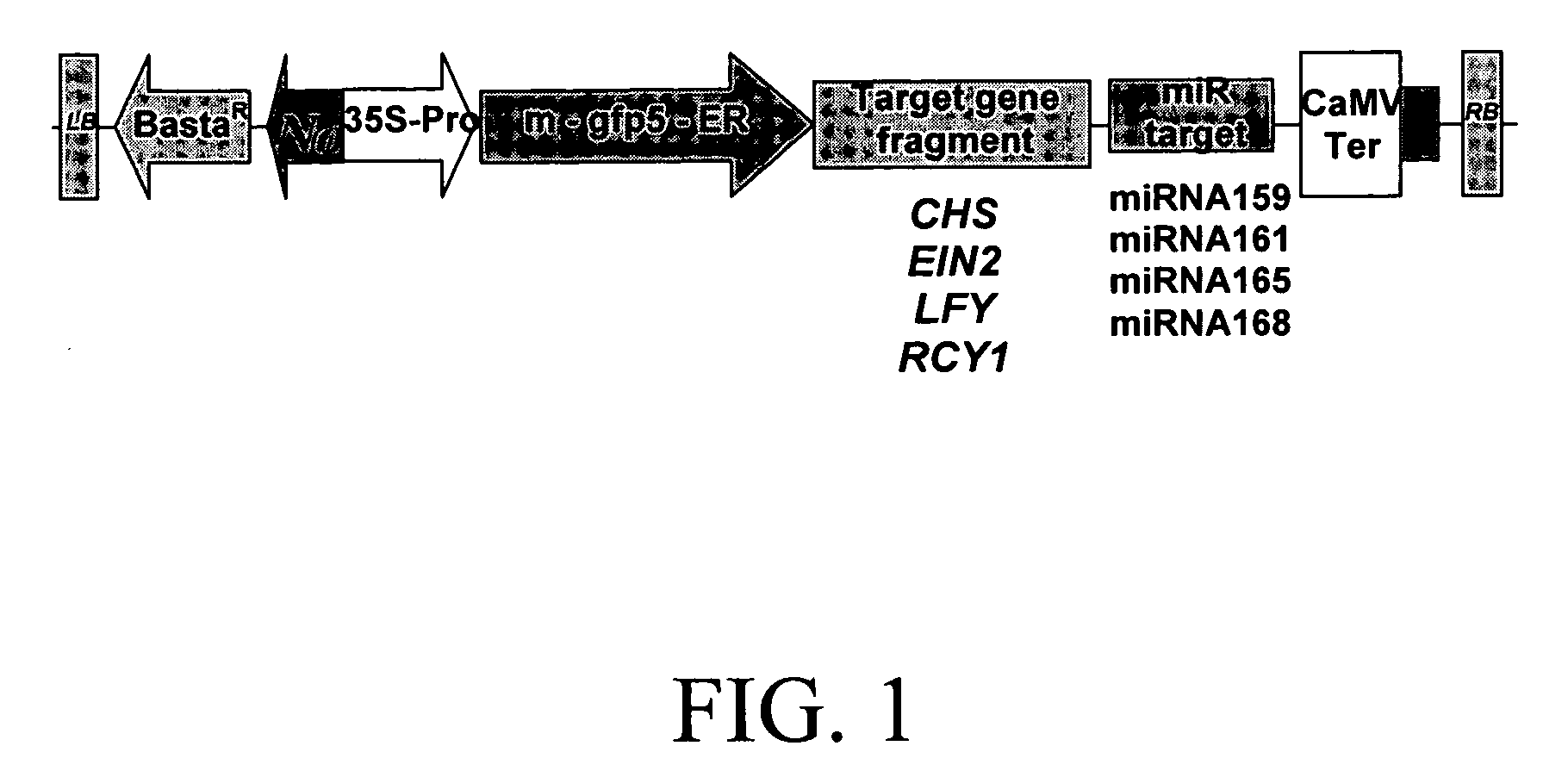
Methods and compositions are provided for reducing the level of expression of a target polynucleotide in an organism. The methods and compositions selectively silence the target polynucleotide through the expression of a chimeric polynucleotide comprising the target for a sRNA (the trigger sequence) operably linked to a sequence corresponding to all or part of the gene or genes to be silenced. In this manner, the final target of silencing is an endogenous gene in the organism in which the chimeric polynucleotide is expressed. In a further embodiment, the miRNA target is that of a heterologous miRNA or siRNA, the latter of which is coexpressed in the cells at the appropriate developmental stage to provide silencing of the final target when and where desired. In a further embodiment, the final target may be a gene in a second organism, such as a plant pest, that feeds upon the organism containing the chimeric gene or genes. Compositions further comprise vectors, seeds, grain, cells, and organisms, including plants and plant cells, comprising the chimeric polynucleotide of the invention.
Owner:PIONEER HI BRED INT INC +1
Pharmaceutical Composition Comprising Anti-Mirna Antisense Oligonucleotides
ActiveUS20100286234A1Reduced microRNA levelAvoid off-target effectsOrganic active ingredientsSugar derivativesNucleobaseRNA Cleavage
The invention provides pharmaceutical compositions comprising short single stranded oligonucleotides, of length of between 8 and 26 nucleobases which are complementary to human microRNAs selected from the group consisting of miR19b, miR21, miR122a, miR155 and miR375. The short oligonucleotides are particularly effective at alleviating miRNA repression in vivo. It is found that the incorporation of high affinity nucleotide analogues into the oligonucleotides results in highly effective anti-microRNA molecules which appear to function via the formation of almost irreversible duplexes with the miRNA target, rather than RNA cleavage based mechanisms, such as mechanisms associated with RNaseH or RISC.
Owner:ROCHE INNOVATION CENT COPENHAGEN
Pharmaceutical composition comprising anti-mirna antisense oligonucleotide
ActiveUS8163708B2Effectively repressedHigh affinitySugar derivativesMetabolism disorderNucleobaseRNA Cleavage
Owner:ROCHE INNOVATION CENT COPENHAGEN
Application of an Epstein-Barr virus miRNA associated with nasopharyngeal carcinoma
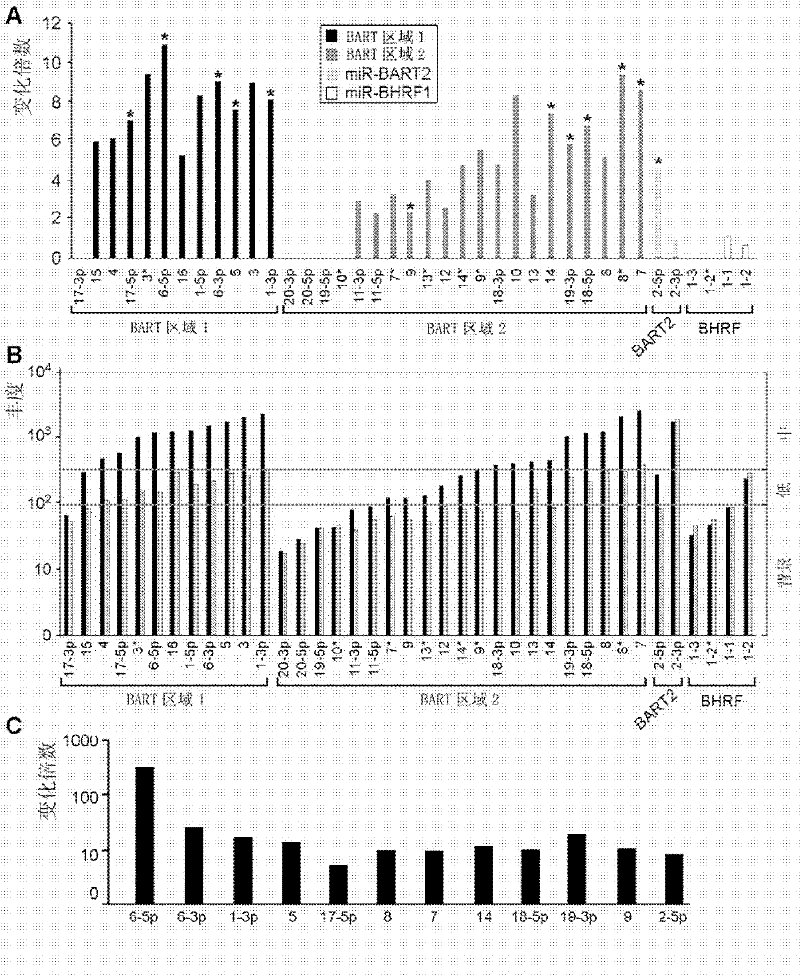
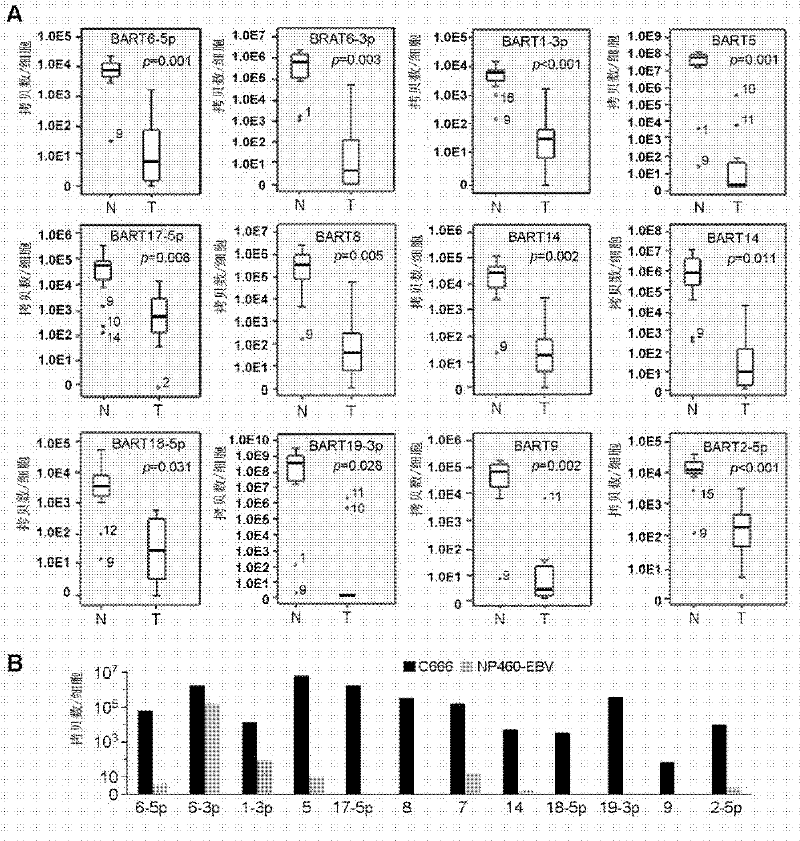
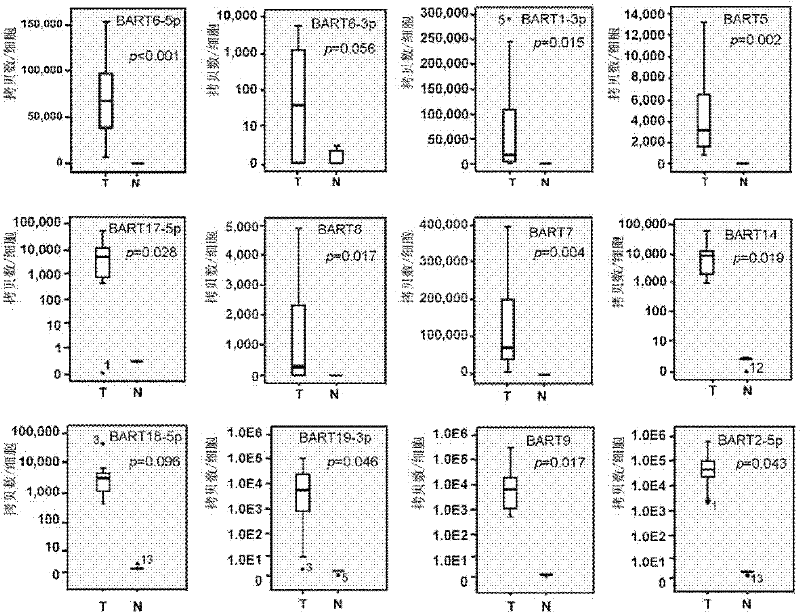
The present invention provides an application of Epstein-Barr virus miRNA related to nasopharyngeal carcinoma, mainly the application of said Epstein-Barr virus miRNA in preparing a kit for diagnosis, prognosis judgment and treatment effect evaluation of nasopharyngeal carcinoma. The present invention also provides a kit for nasopharyngeal carcinoma diagnosis, prognosis judgment, and treatment effect evaluation, said kit comprising Epstein-Barr virus miRNA related to nasopharyngeal carcinoma. The Epstein-Barr virus miRNA related to nasopharyngeal carcinoma includes one or more sequences such as SEQ ID NO: 1-12. The 12 kinds of Epstein-Barr virus miRNAs provided by the present invention can be used for early diagnosis, prognosis judgment, treatment effect evaluation of nasopharyngeal cancer patients, and detection and follow-up of nasopharyngeal cancer high-risk groups by testing the expression level of Epstein-Barr virus miRNAs in serum. At the same time, the study of the target genes of these miRNAs can also provide evidence for Epstein-Barr virus tumorigenesis.
Owner:SUN YAT SEN UNIV CANCER CENT
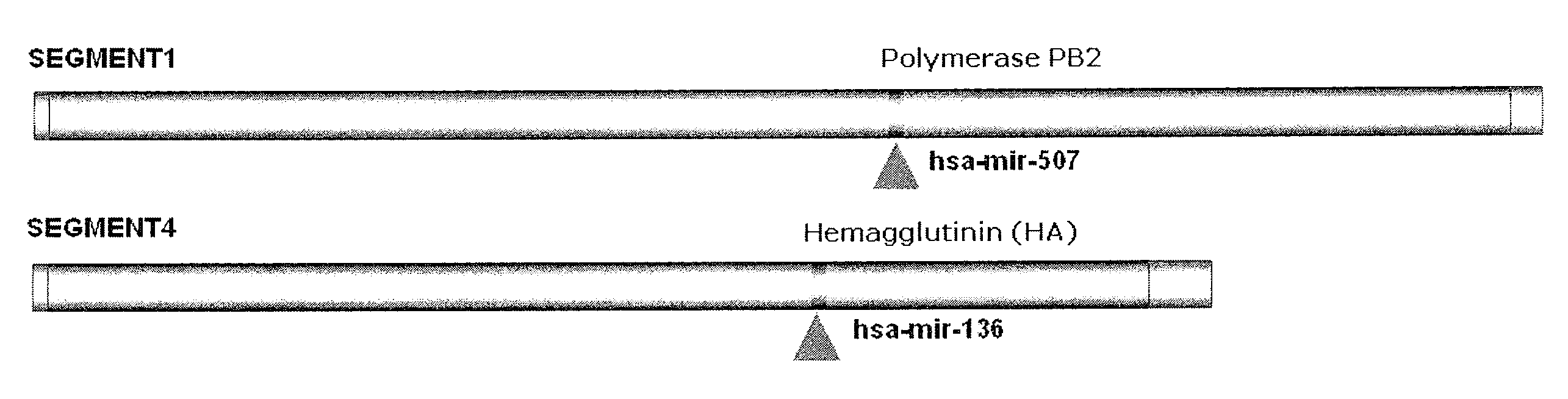
Targets for human micro rnas in avian influenza virus (H5N1) genome
The present invention relates to targets for Human microRNAs in Avian Influenza Virus (H5N1) Genome and provides specific miRNA targets against H5N1 virus. Existing therapies for Avian flu are of limited use primarily due to genetic re-assortment of the viral genome, generating novel proteins, and thus escaping immune response. In animal models, baculovirus-derived recombinant H5 vaccines were immunogenic and protective, but results in humans were disappointing even when using high doses. Currently, two classes of drugs are available with antiviral activity against influenza viruses: inhibitors of the M2 ion channel, amantadine and rimantadine, and inhibitors of neuraminidase, oseltamivir, and zanamivir. There is paucity of information regarding effectiveness of these drugs in H5N1 infection. These drugs are also well known to have side effects like neurotoxicity. Thus there exists a need to develop alternate therapy for targeting the Avian flu virus (H5N1). The present invention addresses this need in the field.
Owner:COUNCIL OF SCI & IND RES
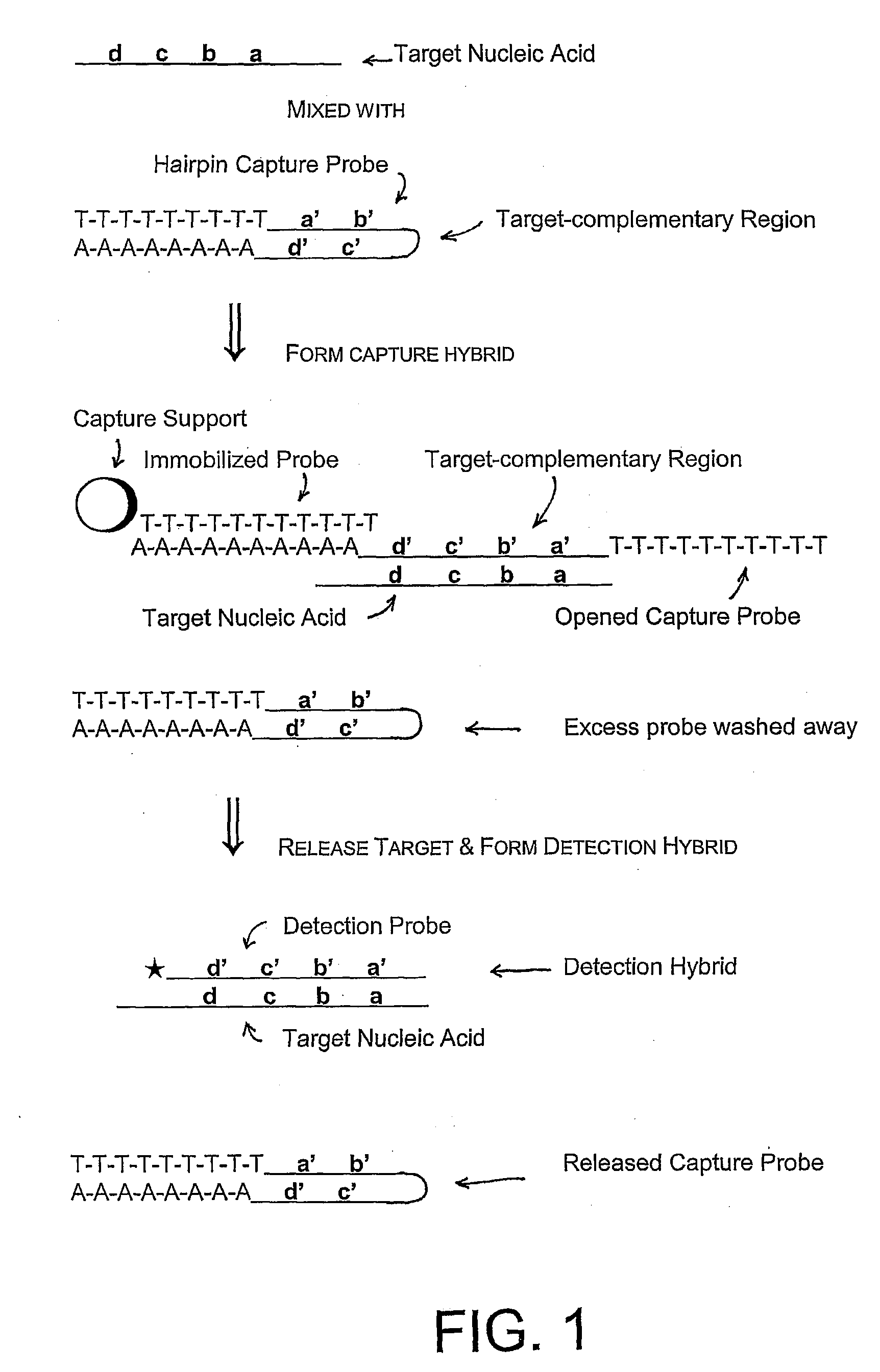
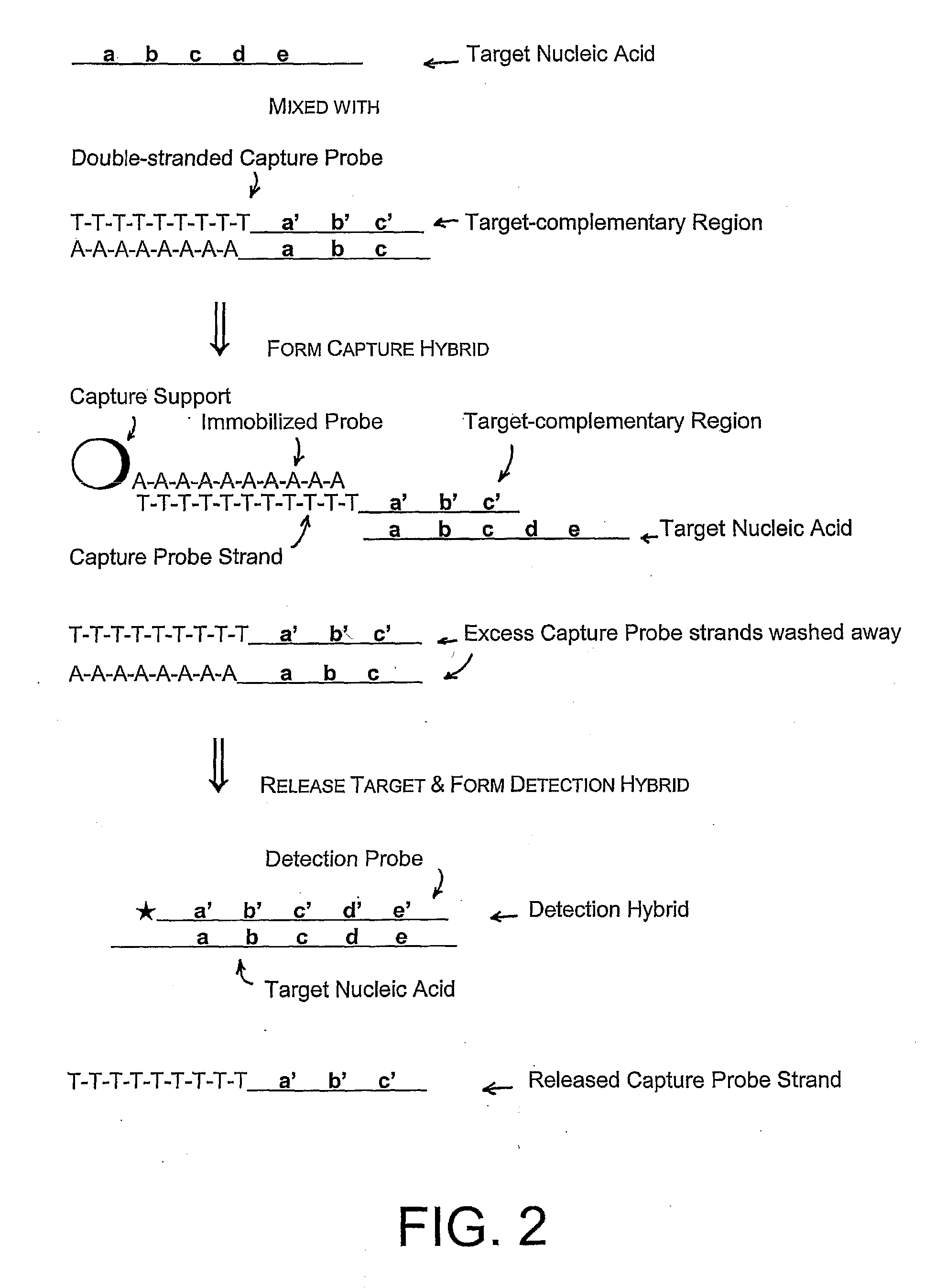
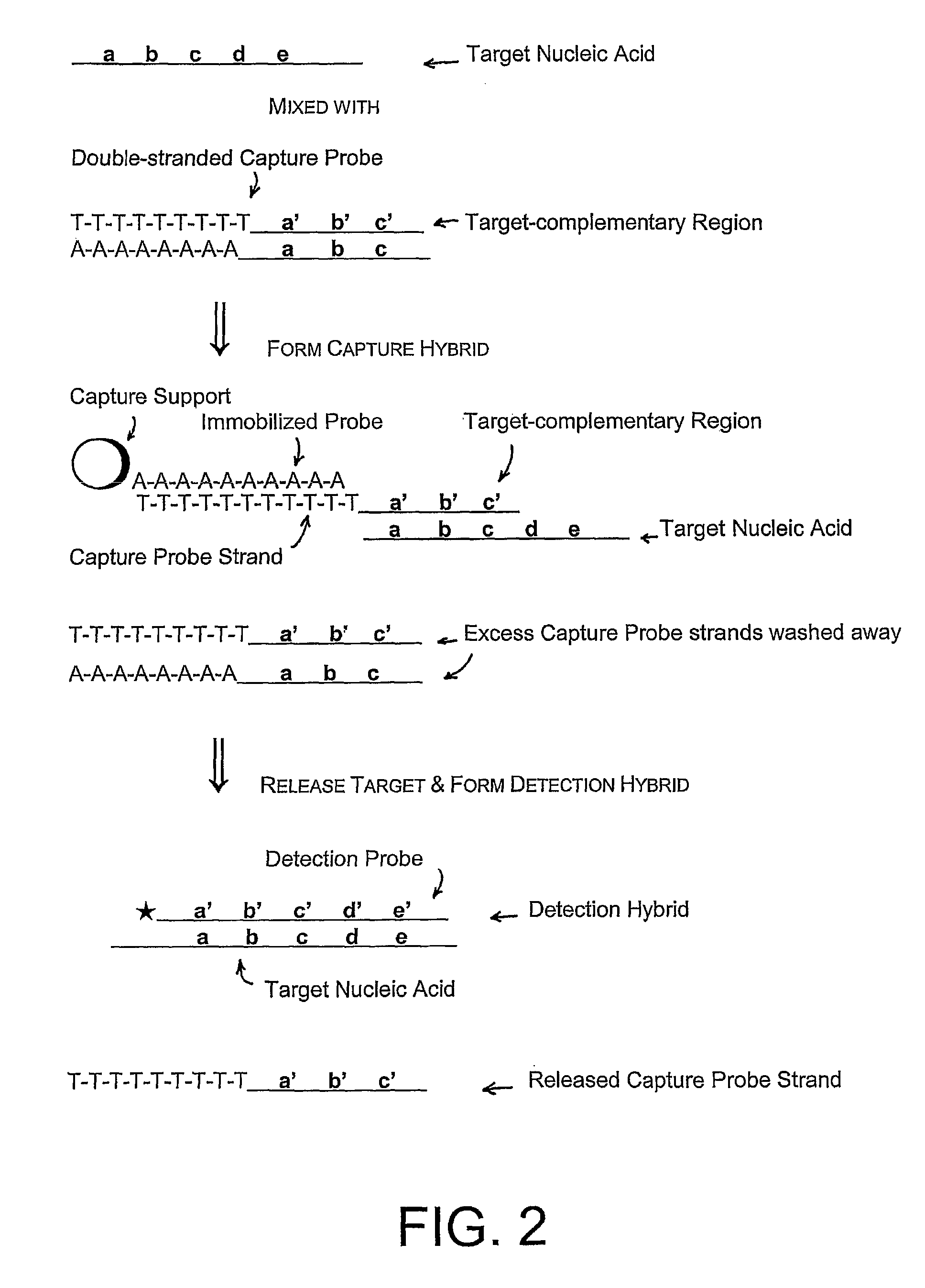
Compositions and methods for detecting small rnas, and uses thereof
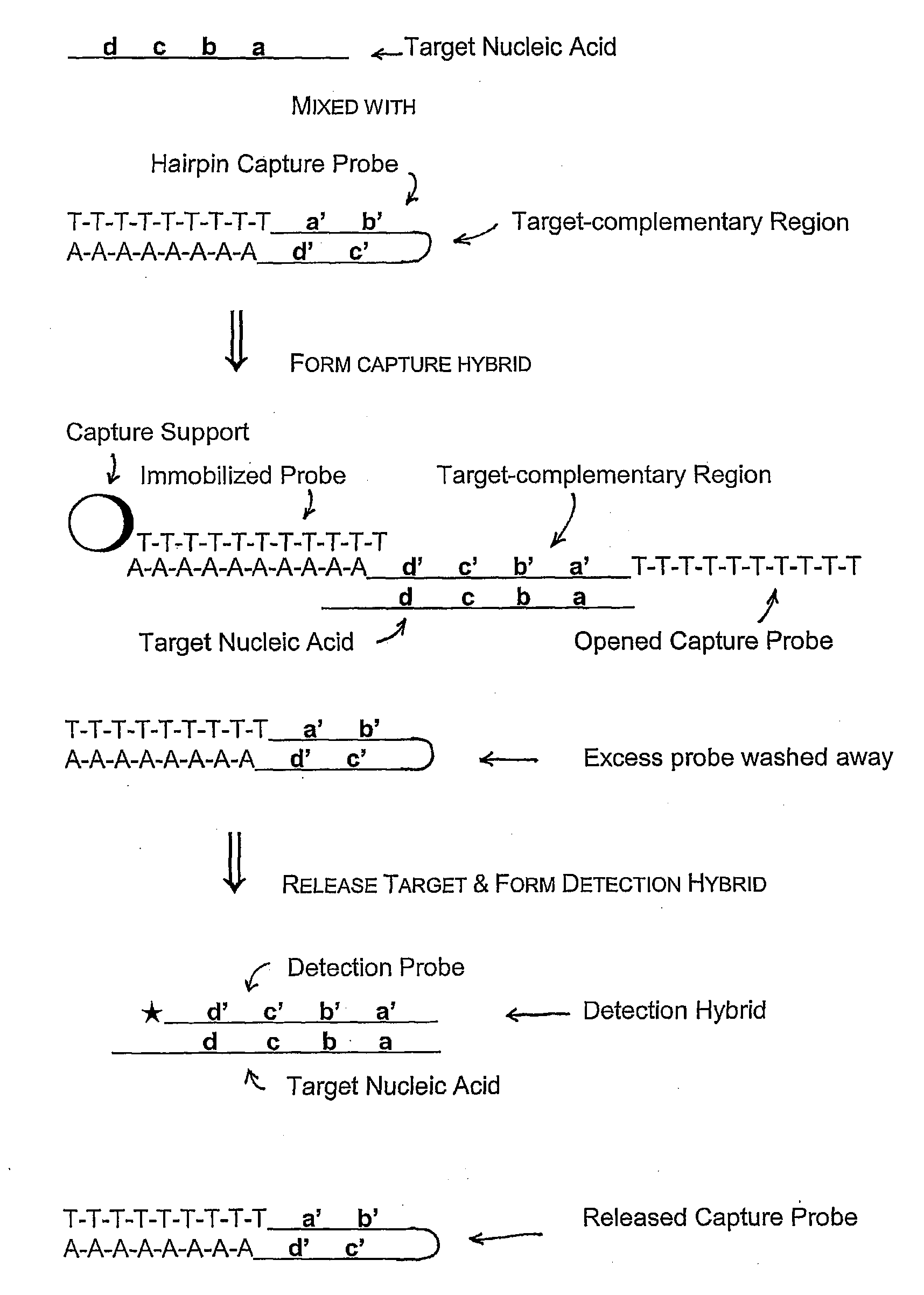
Compositions and methods are provided for the detection of small RNA target nucleic acids, preferably miRNA target nucleic acids, wherein the compositions and methods provide for sensitive and specific detection of the target nucleic acids. The compositions and methods include using one or more of a first amplification oligomer that is preferably an extender primer, a target capture oligomer that is preferably at least partially double stranded, a promoter primer / provider, a reverse primer that is preferably a universal primer and a detection probe. The compositions and methods are useful for diagnostics, prognostics, monitoring the effectiveness of treatment and / or determining a treatment.
Owner:GEN PROBE INC
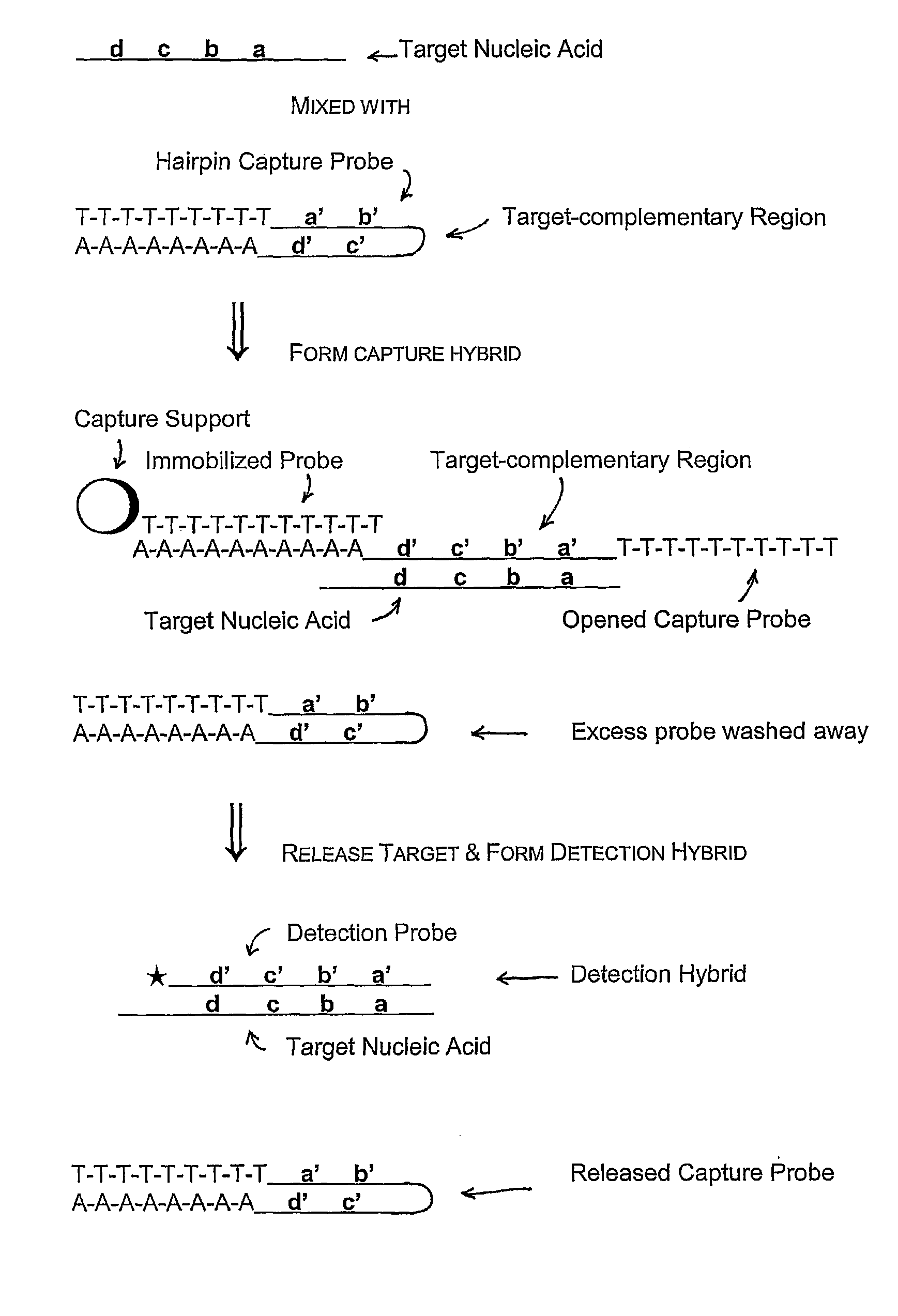
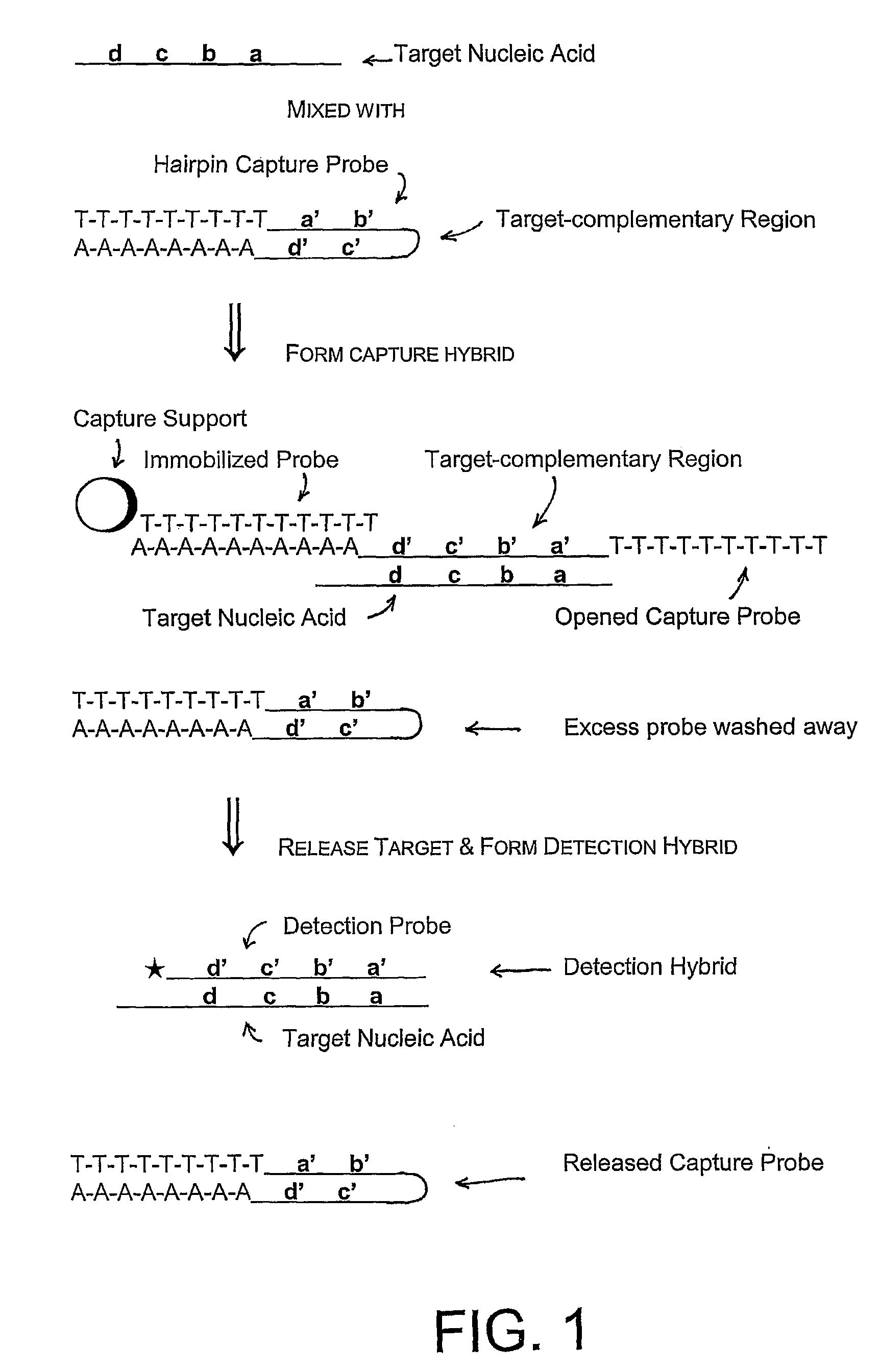
Compositions and methods for detecting small RNAs, and uses thereof
Compositions and methods are provided for the detection of small RNA target nucleic acids, preferably miRNA target nucleic acids, wherein the compositions and methods provide for sensitive and specific detection of the target nucleic acids. The compositions and methods include using one or more of a first amplification oligomer that is preferably an extender primer, a target capture oligomer that is preferably at least partially double stranded, a promoter primer / provider, a reverse primer that is preferably a universal primer and a detection probe. The compositions and methods are useful for diagnostics, prognostics, monitoring the effectiveness of treatment and / or determining a treatment.
Owner:GEN PROBE INC
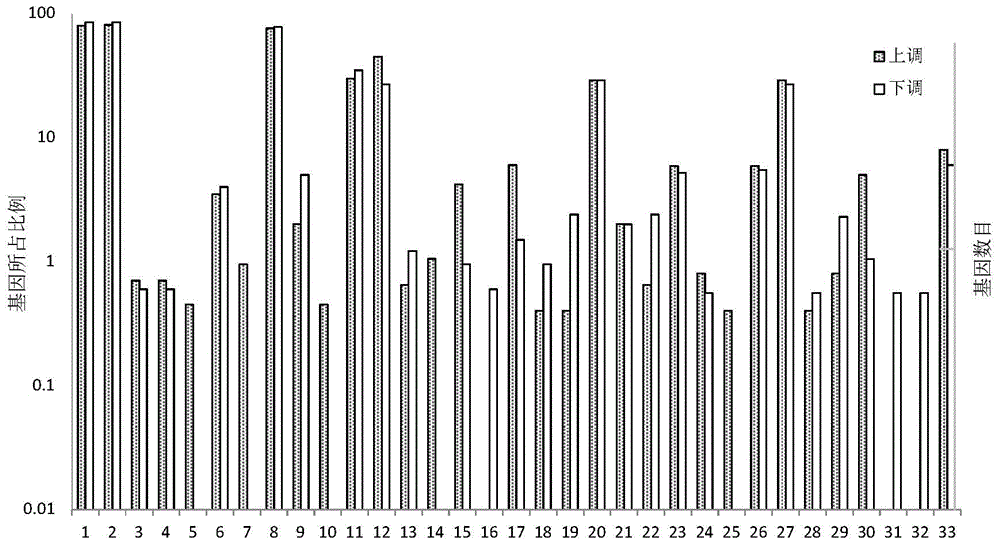
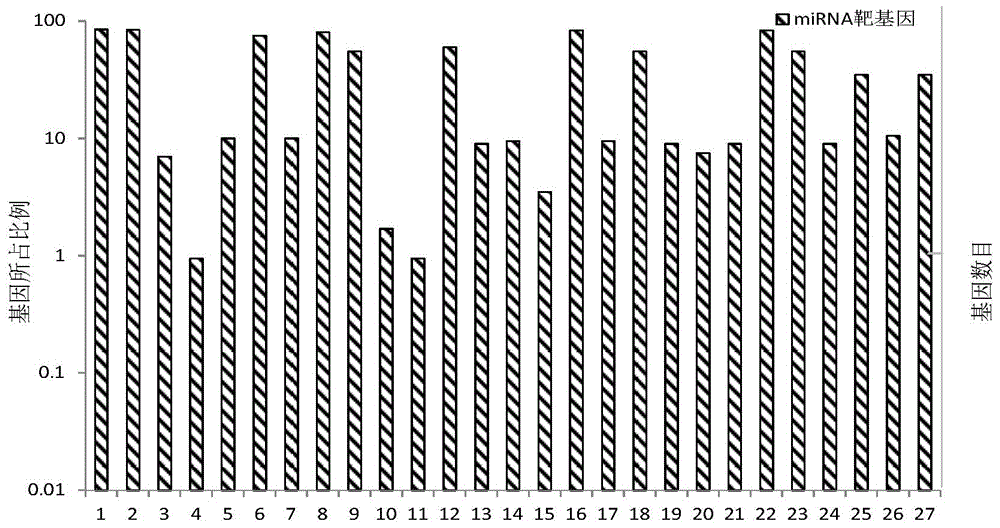
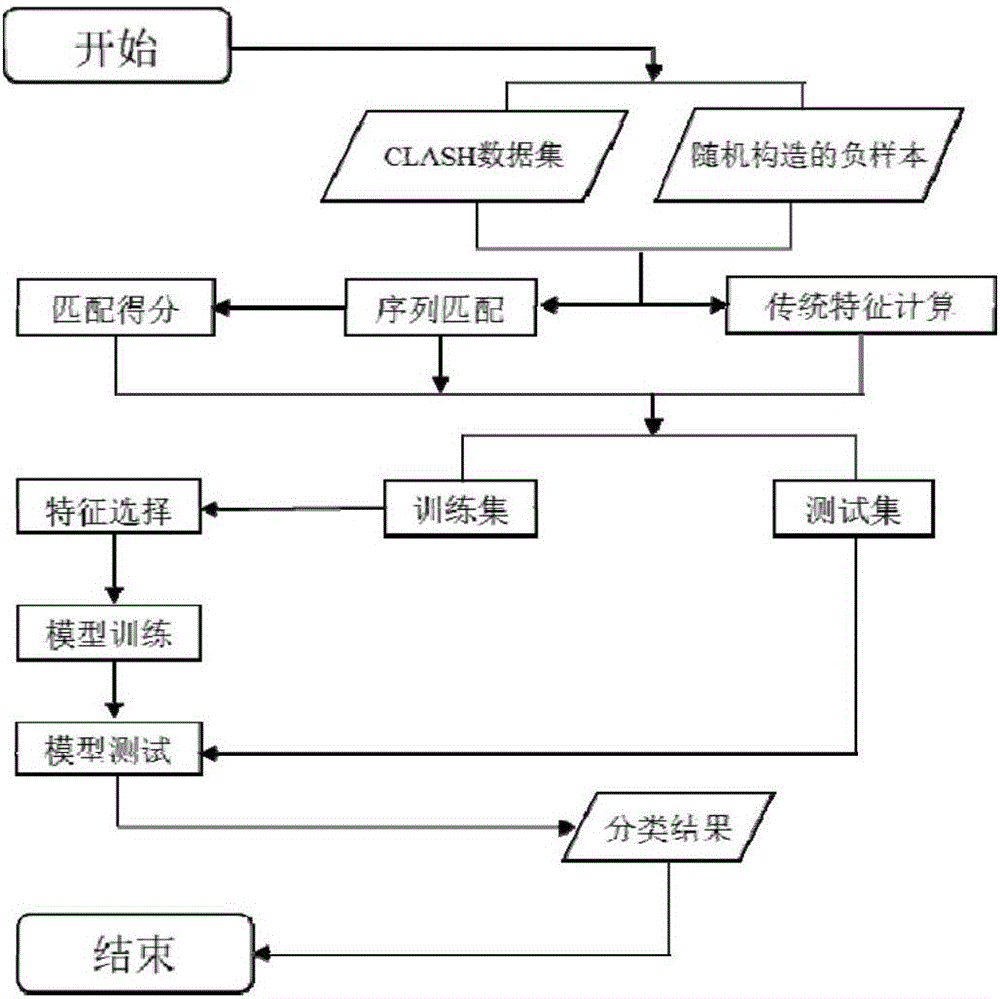
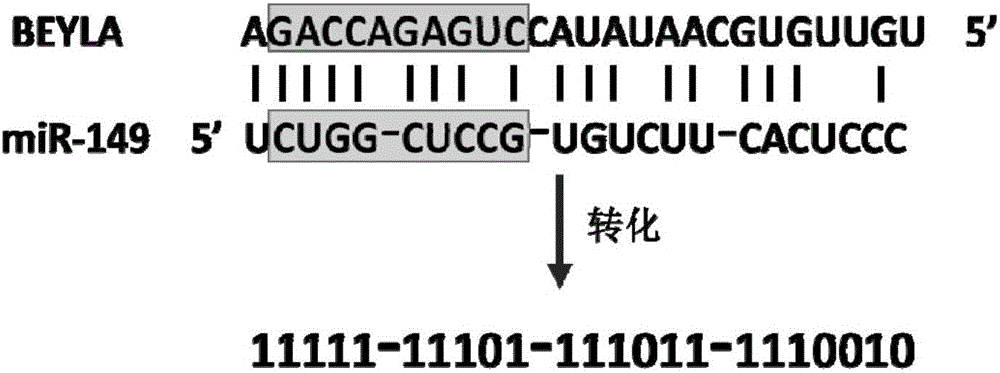
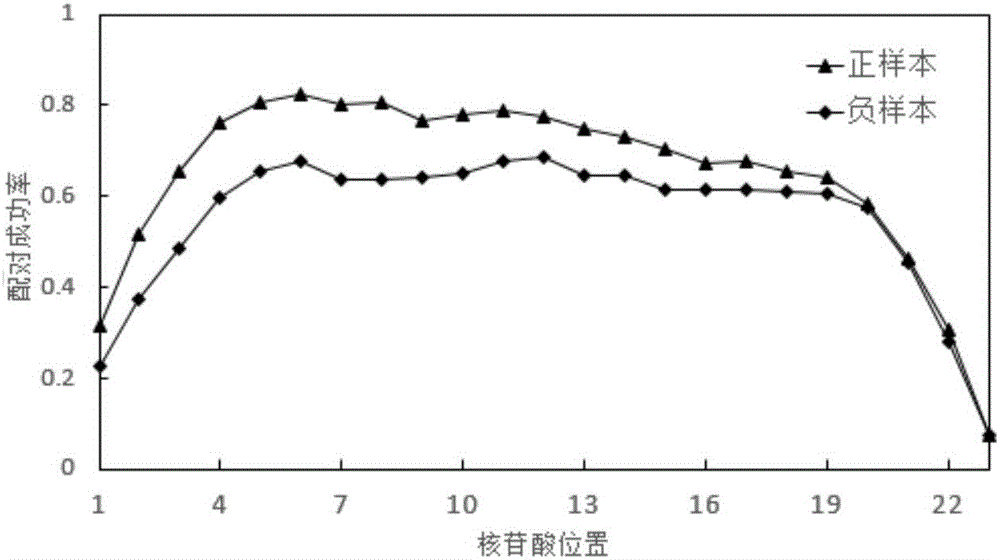
Sequence characteristic analysis method for forecasting miRNA target gene
ActiveCN106599615AMeasuring Binding PossibilitiesBalance differenceBiostatisticsSequence analysisData setRelevant feature
The invention discloses a sequence characteristic analysis method for forecasting a miRNA target gene. The method comprises the steps of constructing related characteristics of 27 miRNA-target point pairing sequences on the basis of a CLASH experiment data set, and forming a characteristic set comprising 84 characteristic values by combining traditional characteristic; and performing machine learning by using a random forest model, and constructing a miRNA target gene forecast model to perform miRNA target gene recognition. The model constructed according to the method has the advantages of high accuracy, sensitivity, specificity and precision, and the miRNA target gene can be relatively and accurately forecasted.
Owner:SYSU CMU SHUNDE INT JOINT RES INST +2
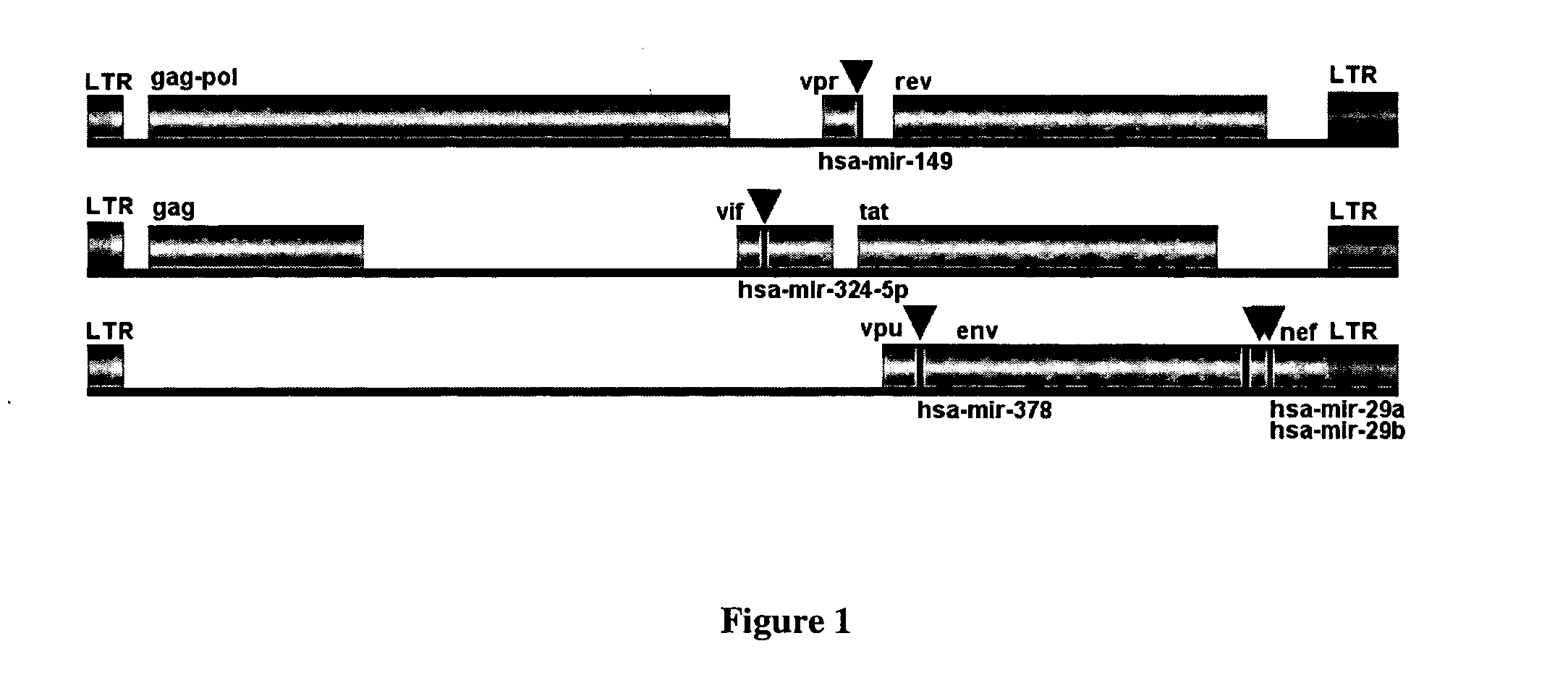
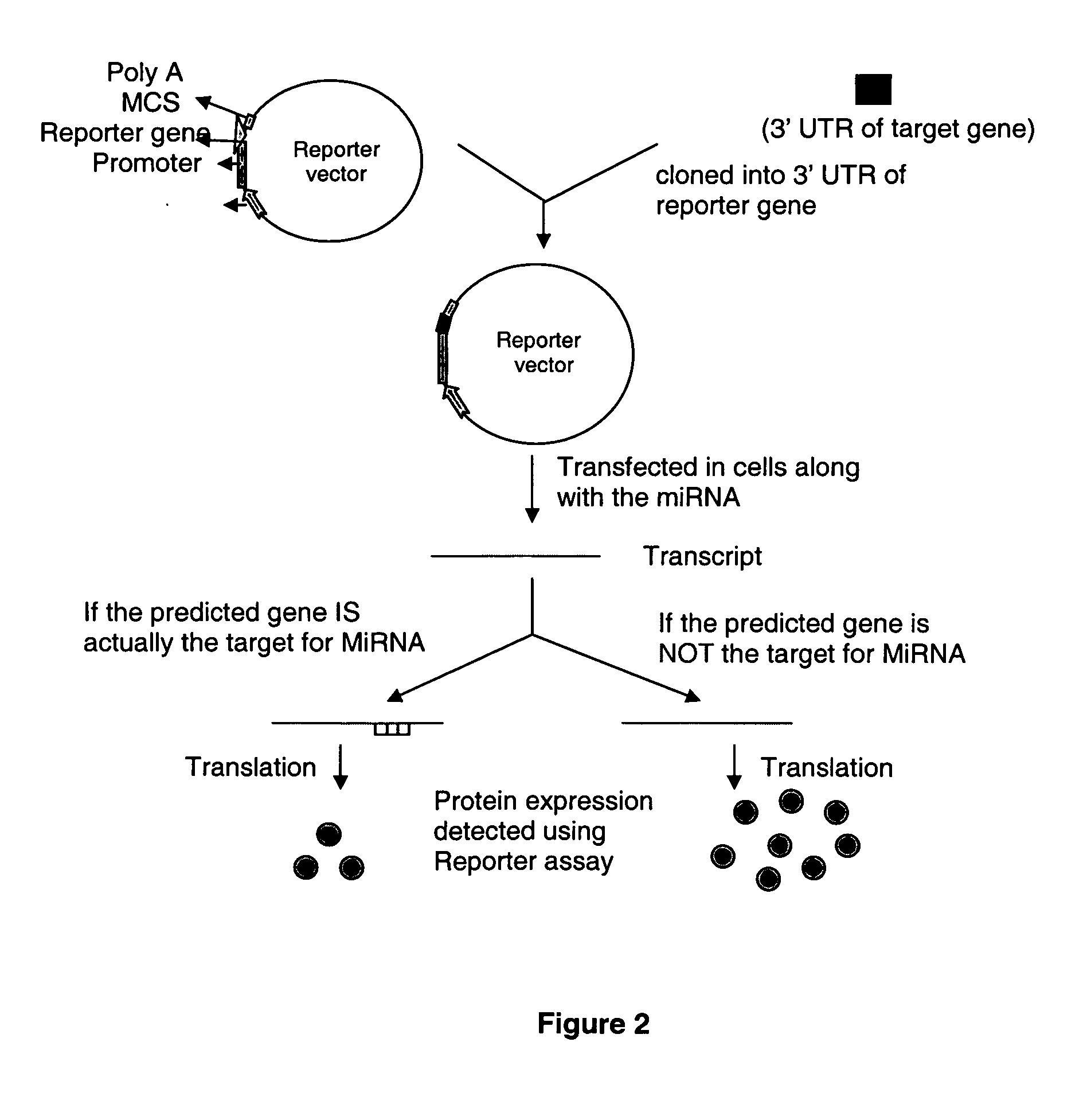
Human microRNA targets in HIV genome and a method of identification thereof
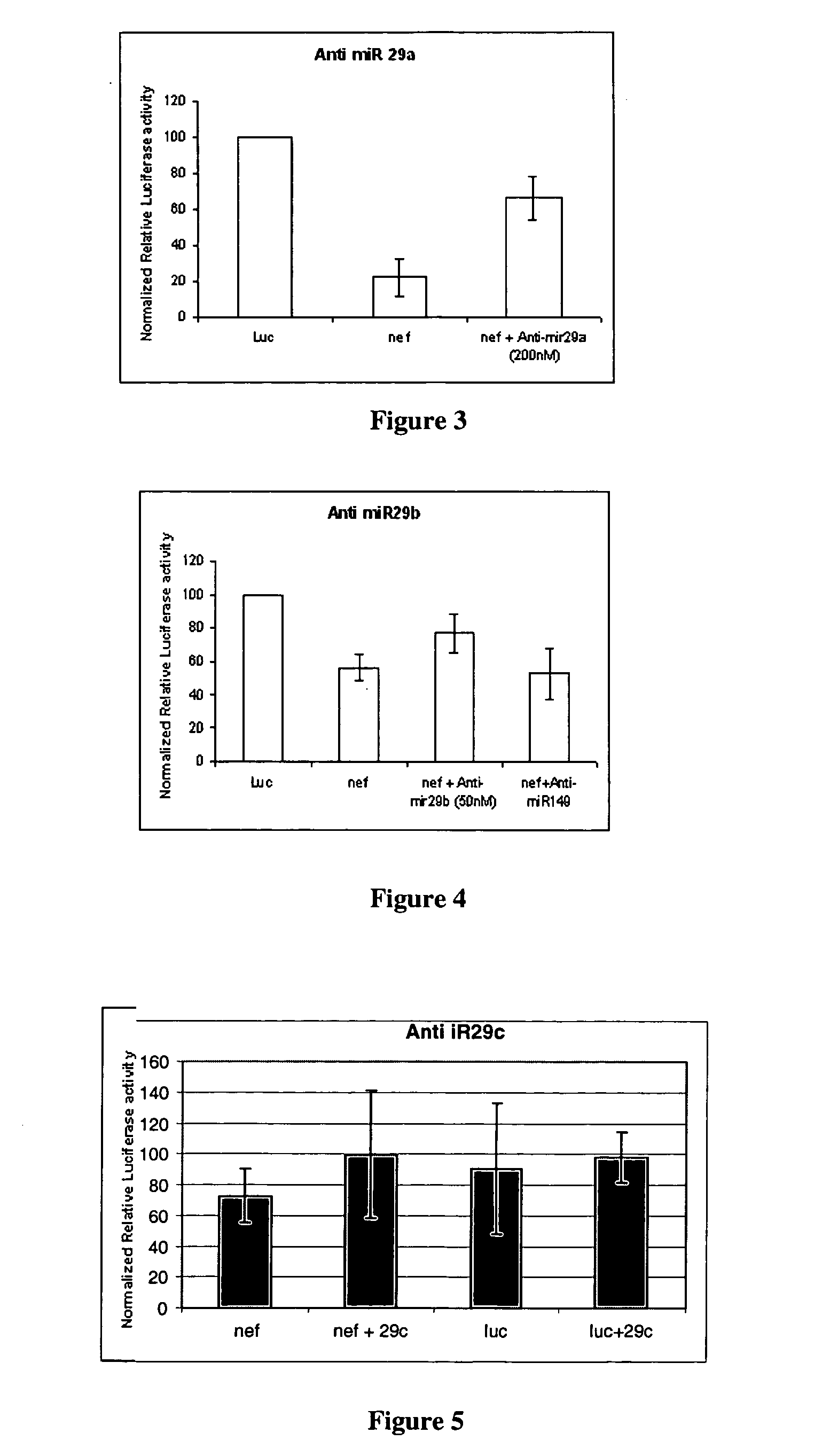
The present invention relates to human microRNA targets in HIV genome and a method of identification thereof. Using multiple software targets to six human microRNAs [miRNAs] were discovered in the net, vpr, env, and I vif genes. The miRNAs were identified as hsa-miR-29a, hsa-miR-29b, hsa-miR-29c, hsa-mir-149, hsa-mir-324-5p, hsa-mir-378. These miRNAs or its homologues can be used as therapeutics against HIV infection. The invention further relates to a novel strategy to target genes of HIV-1 by human microRNAs, or its homologues, to inactivate or block HIV activity. The computational approach towards identification of human miRNA targets in the HIV genome and the variation in the microRNA levels was further validated experimentally.
Owner:COUNCIL OF SCI & IND RES
Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks
InactiveCN106529203AConvenient researchData visualisationSystems biologyProtein targetProtein insertion
The invention discloses a method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks. The method includes steps of building three sub-networks including a human protein-protein interaction network on the basis of HIPPIE, a miRNA-target protein network on the basis of mirTARbase and a miRNA-miRNA network on the basis of target protein overlapping structures; combining the three sub-networks with one another according to acquisition numbers of proteins and ID (identification) numbers of miRNA molecules in miRbase databases and building fusion miRNA-target protein association relationship networks; representing miRNA-target protein association features on the basis of guilt-by-association principles, building classification prediction models by the aid of random forest and predicting potential interaction association relationships between miRNA and the target proteins. The method has the advantages that research on many-to-many relationships of the miRNA regulation target proteins can be effectively carried out, and accordingly the method has high application value.
Owner:SYSU CMU SHUNDE INT JOINT RES INST +2
miRNA Target Prediction
Owner:RGT UNIV OF MICHIGAN
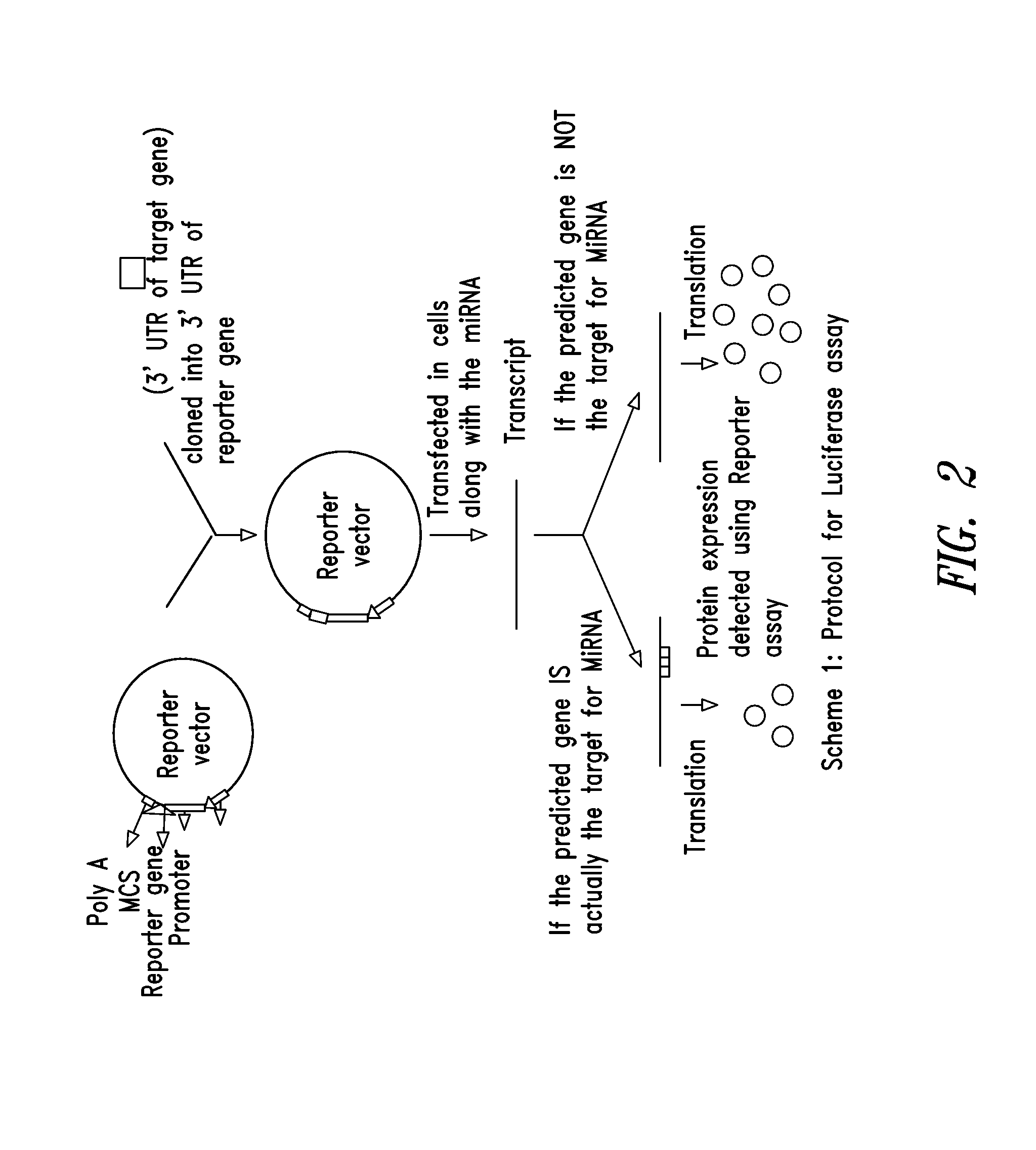
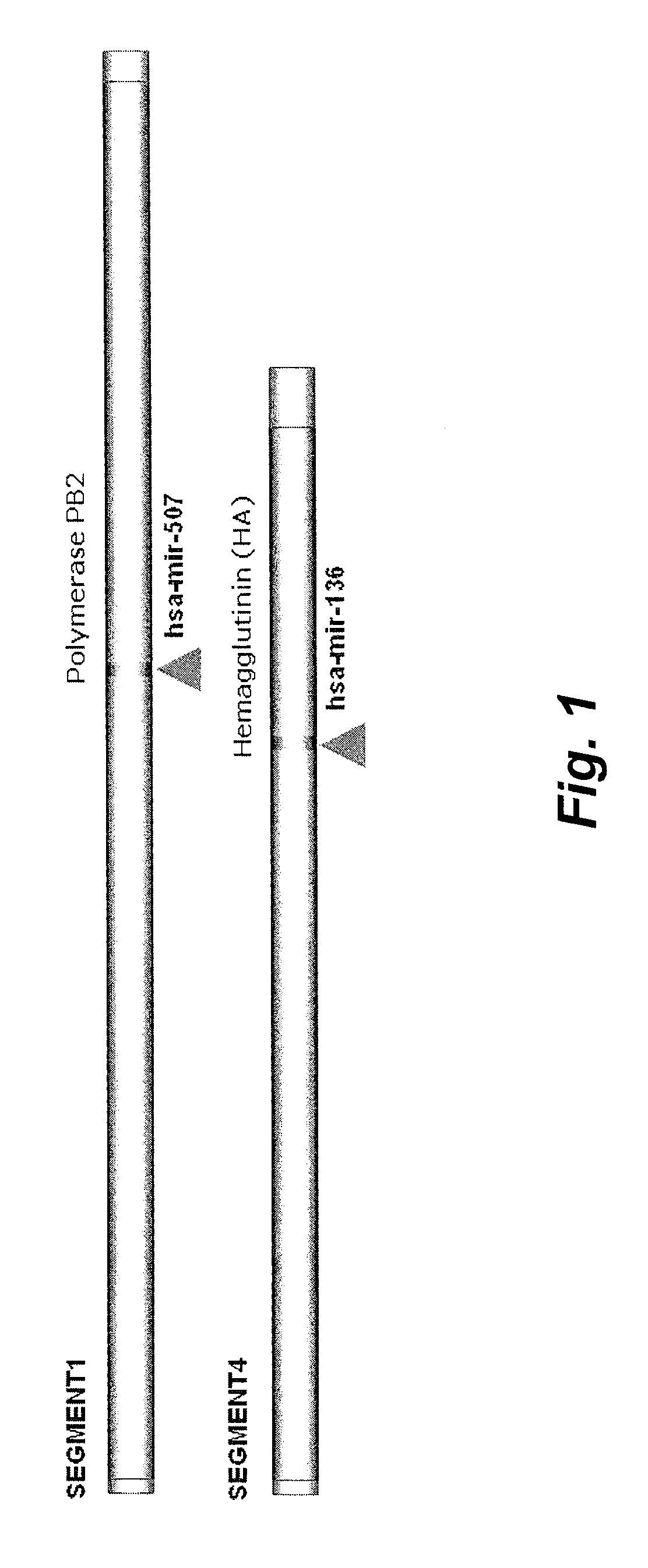
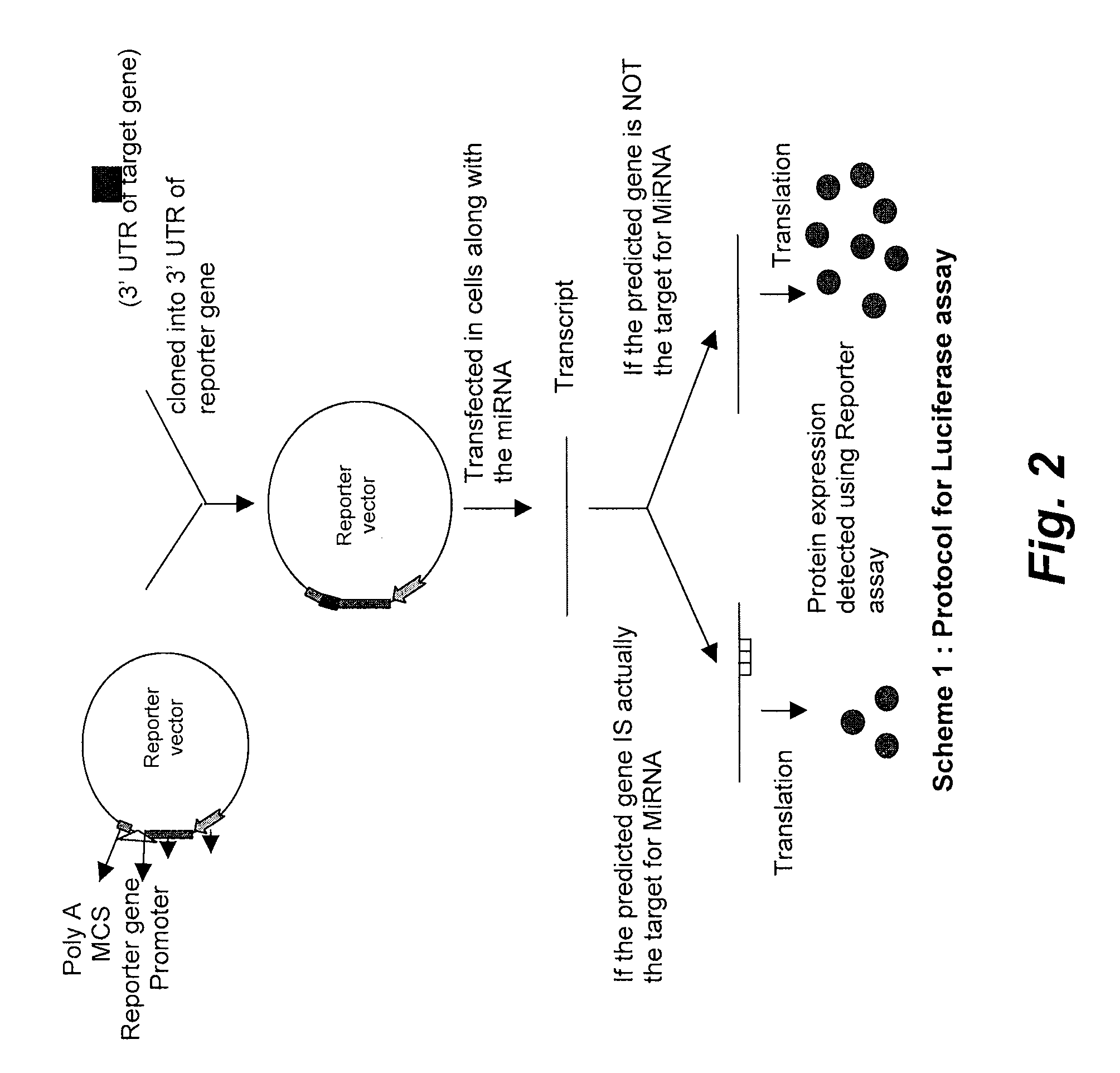
Novel multifunctional dual-luciferase reporter gene plasmid
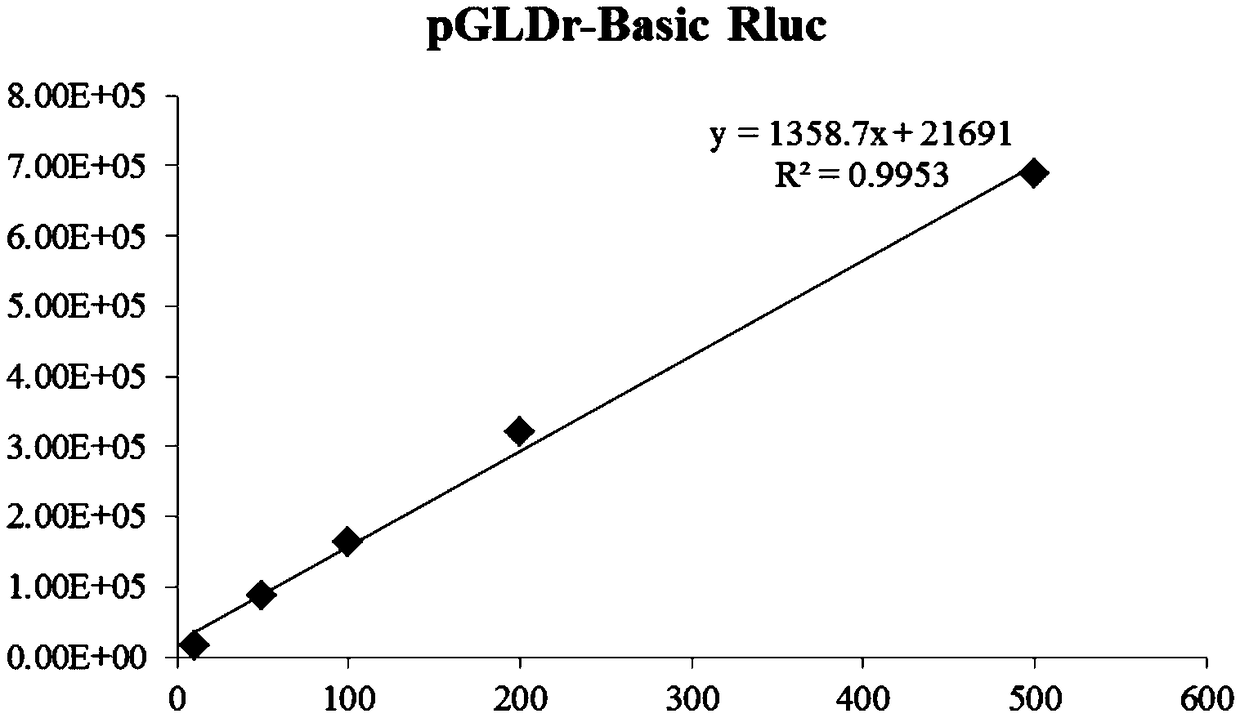
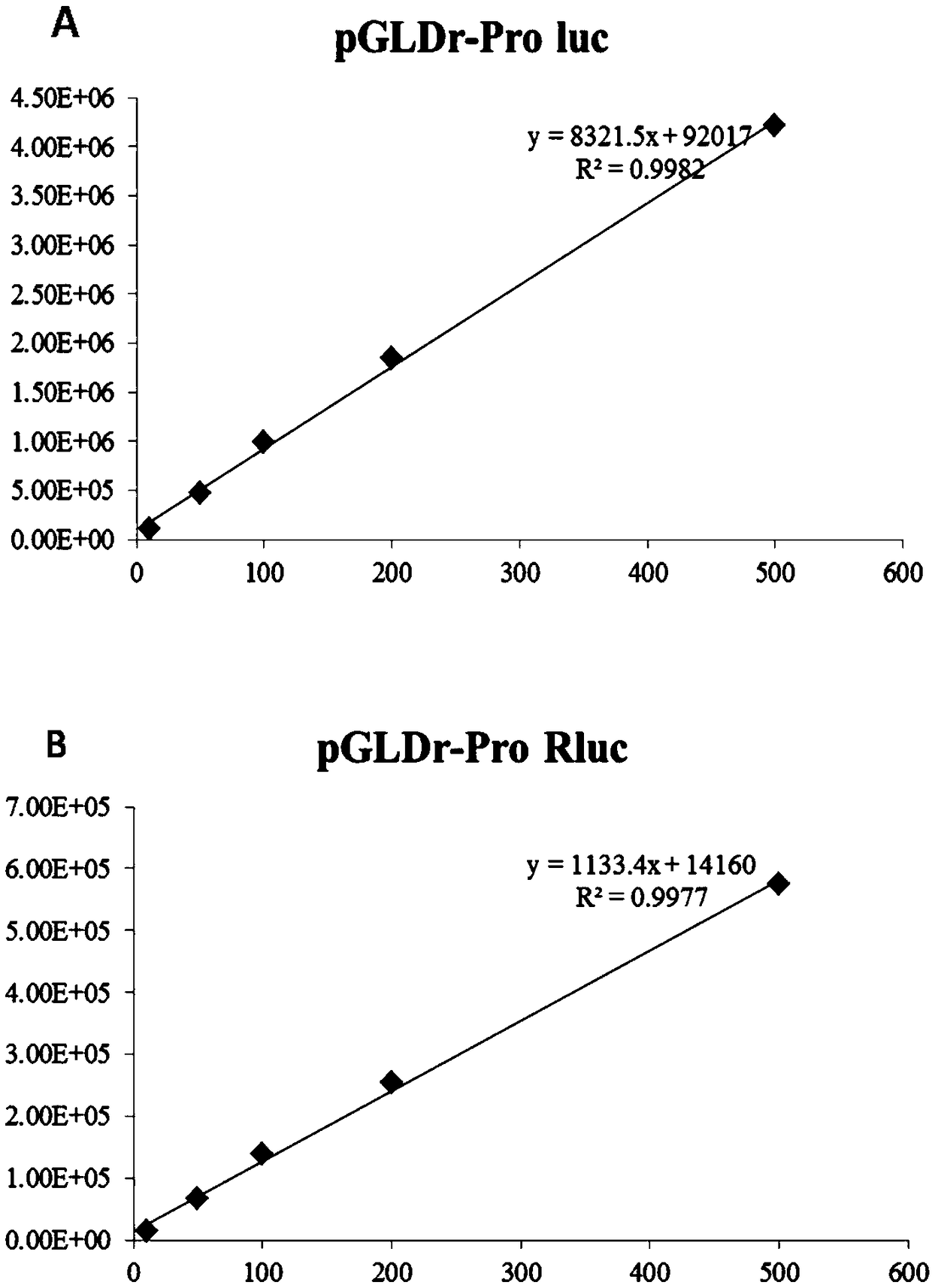
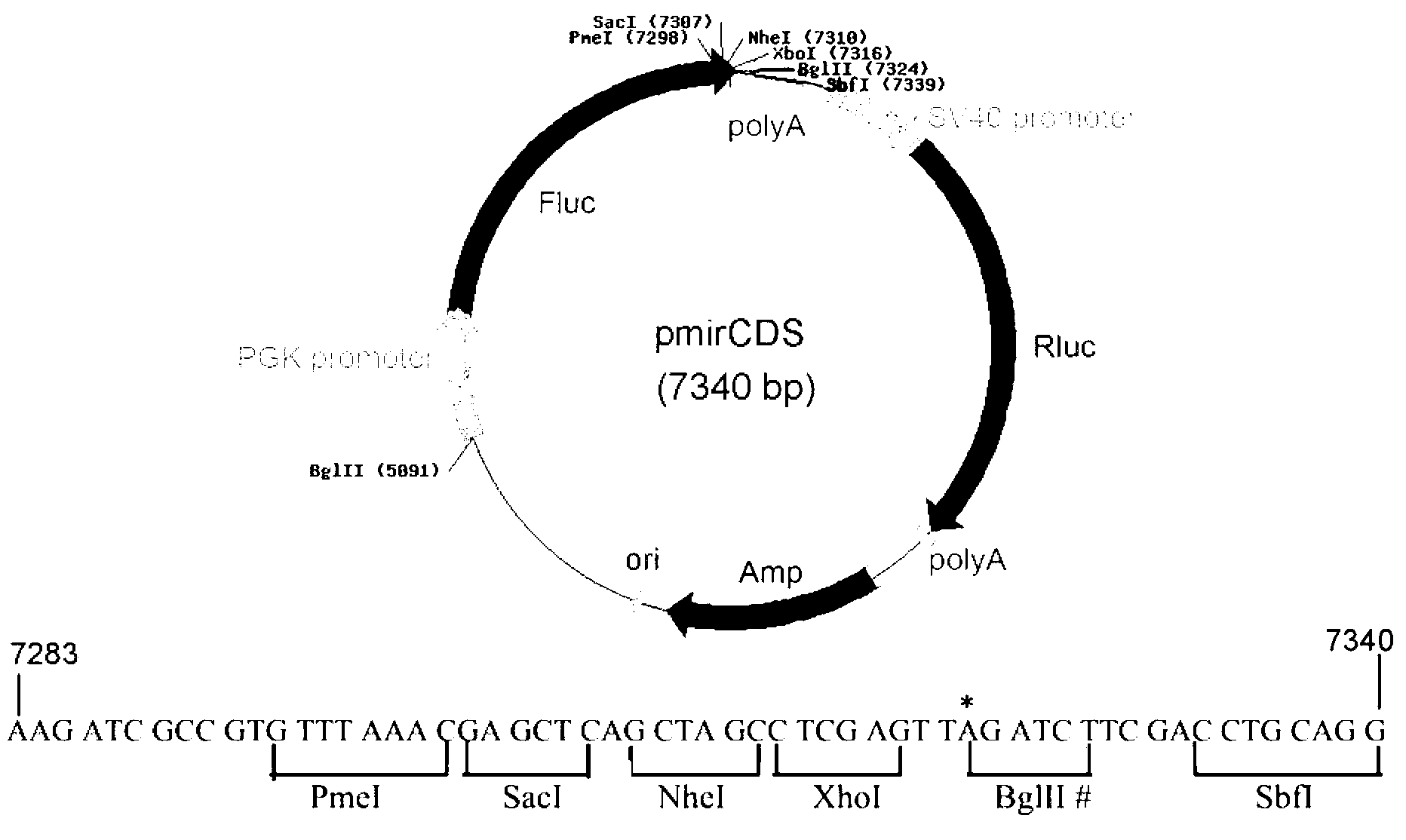
InactiveCN109022467AReduce typesReduce experimental errorOxidoreductasesVector-based foreign material introductionRenilla luciferaseMultiple cloning site
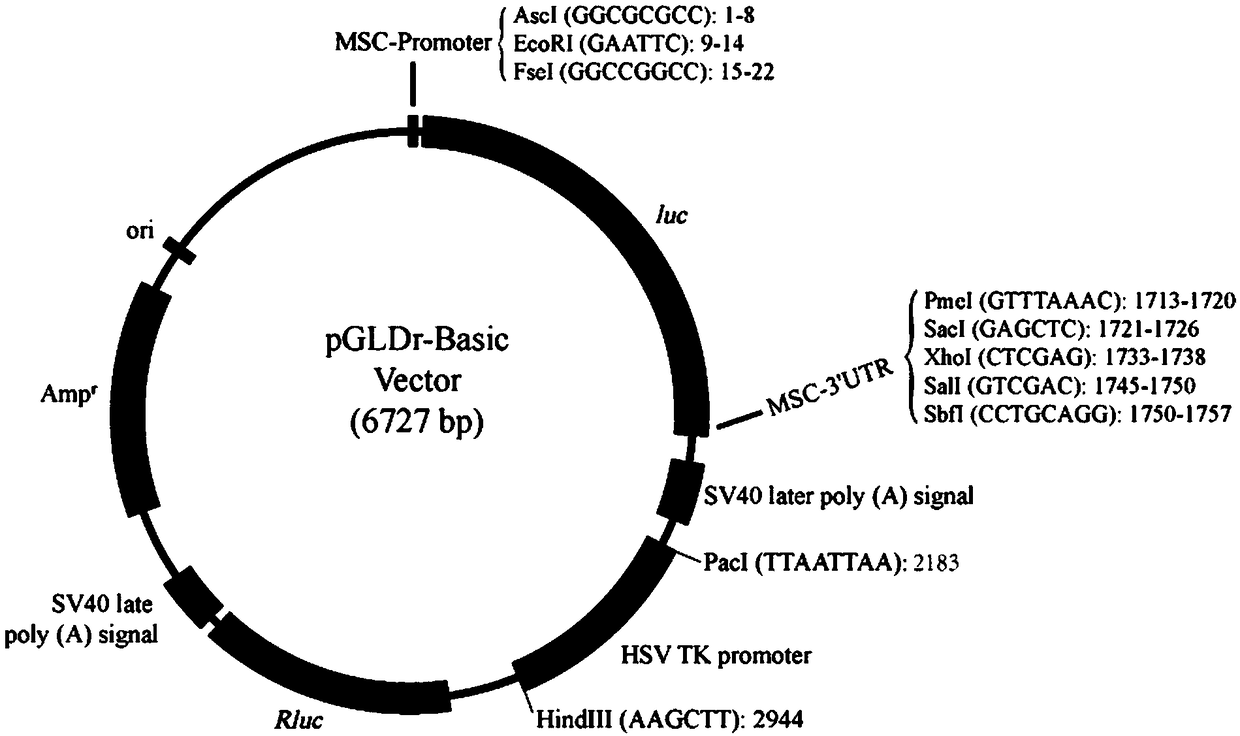
The invention discloses a novel multifunctional dual-luciferase reporter gene plasmid. The plasmid contains a luc firefly luciferase gene, a Rluc renilla luciferase gene, a SV40 poly(A) termination signal part and two independent multiple cloning sites, wherein the two independent multiple cloning sites are located on the upstream and the downstream of a luc firefly luciferase gene coder frame respectively. The dual-luciferase reporter gene plasmid containing detection gene and reference gene is successfully constructed, not only can the detection gene be transferred into a subject cell conveniently, but also the reference gene can be taken in correspondingly, the complexity of experiment operation and experiment errors are greatly reduced, the plasmid has the multiple cloning sites at thetwo ends of the firefly luciferase gene, the multiple cloning site located on the upstream of firefly luciferase can introduce a promoter sequence which can be used for genetic transcription adjustment, control and detection, and the multiple cloning site located on the downstream of the firefly luciferase can introduce a 3'UTR sequence which can be used for miRNA target identification, so that switching of different application of the same plasmid is achieved.
Owner:SUN YAT SEN UNIV
Dual fluorescent reporter gene vector for identifying miRNA targets, preparation method and application thereof
InactiveCN103266120AEasy to operateShort experiment cycleMicrobiological testing/measurementVector-based foreign material introductionRenilla luciferaseMultiple cloning site
The invention discloses a dual fluorescent reporter gene vector for identifying miRNA targets, a preparation method thereof, and an application thereof in identifying the miRNA targets in coding regions and detecting functions of the miRNA on the coding regions. The carrier has a renilla luciferase gene and a firefly luciferase gene, and multiple cloning sites (MCS) are located between a terminal amino acid codon and a stop codon of a firefly luciferase reporter gene, thereby facilitating insertion of the miRNA targets into the coding regions of the firefly luciferase reporter gene, so as to identify the miRNA targets in coding regions and detect the functions of the miRNA on the coding regions. The preparation method of the carrier provided by the invention does not require a special treatment for terminals of the carrier or target fragments, is free of connection reaction and PCR amplification, and is simple to operate, short in experiment period and high in efficiency.
Owner:HUNAN NORMAL UNIVERSITY
Recombinant DNA constructs and methods for modulating expression of a target gene
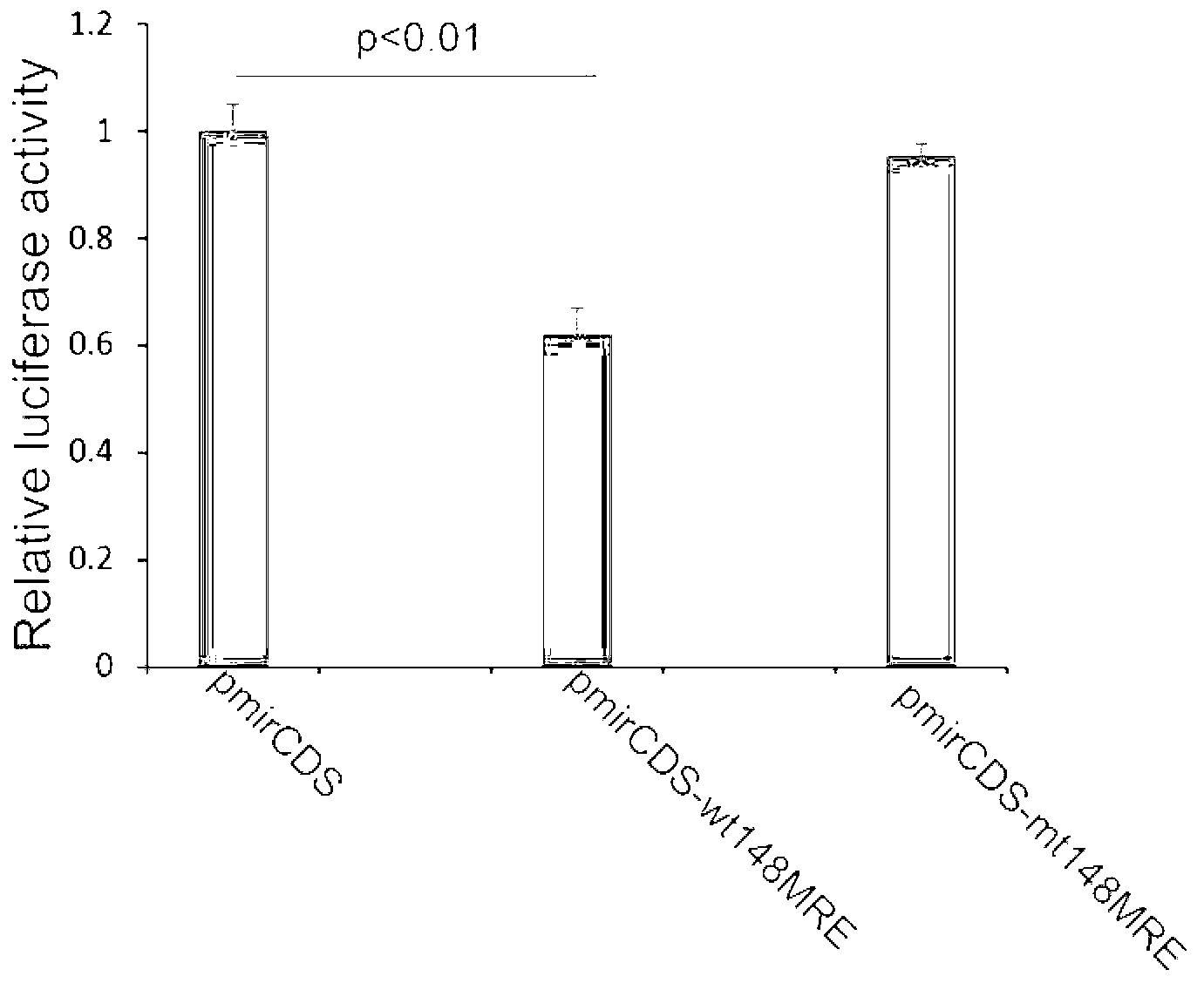
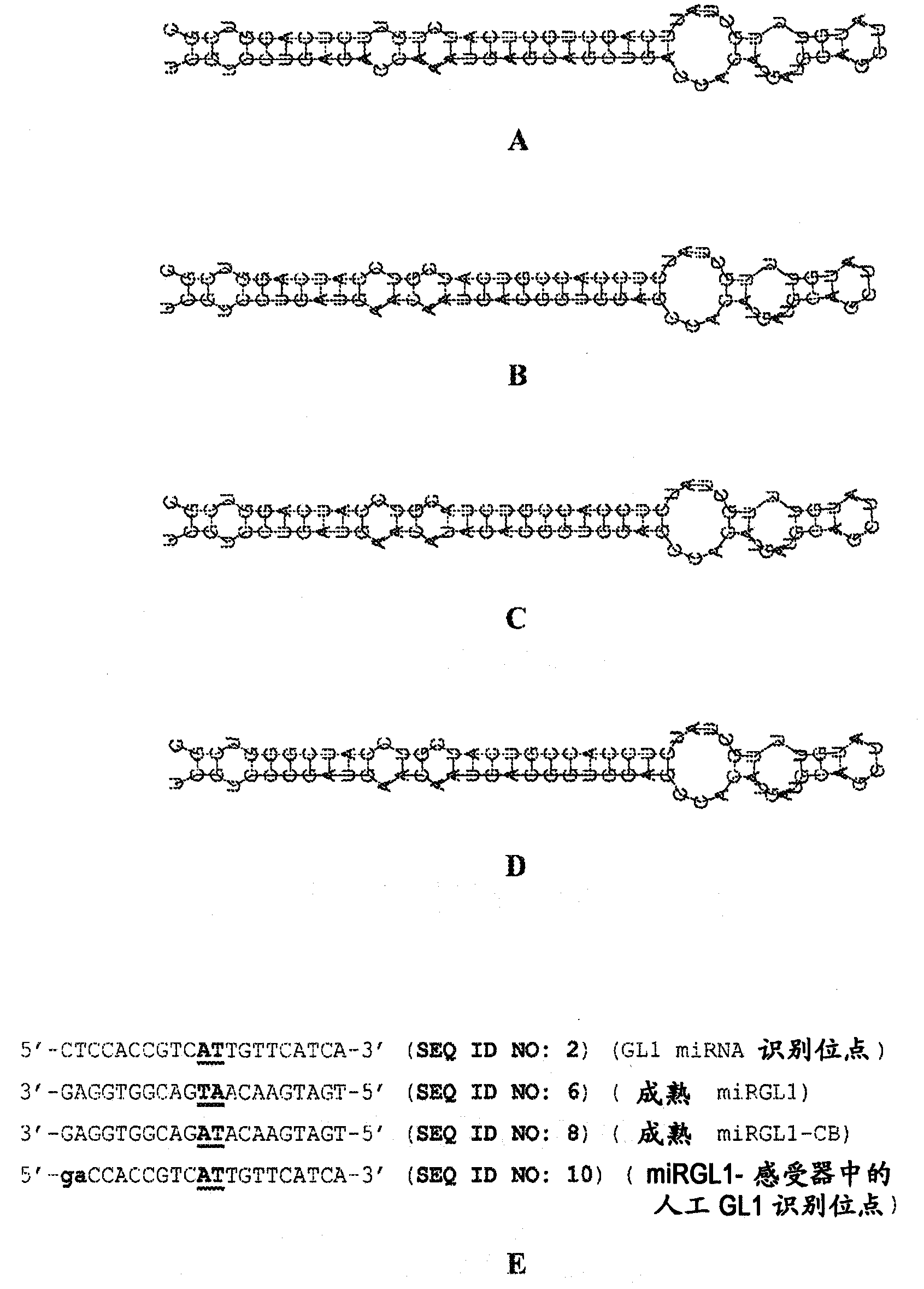
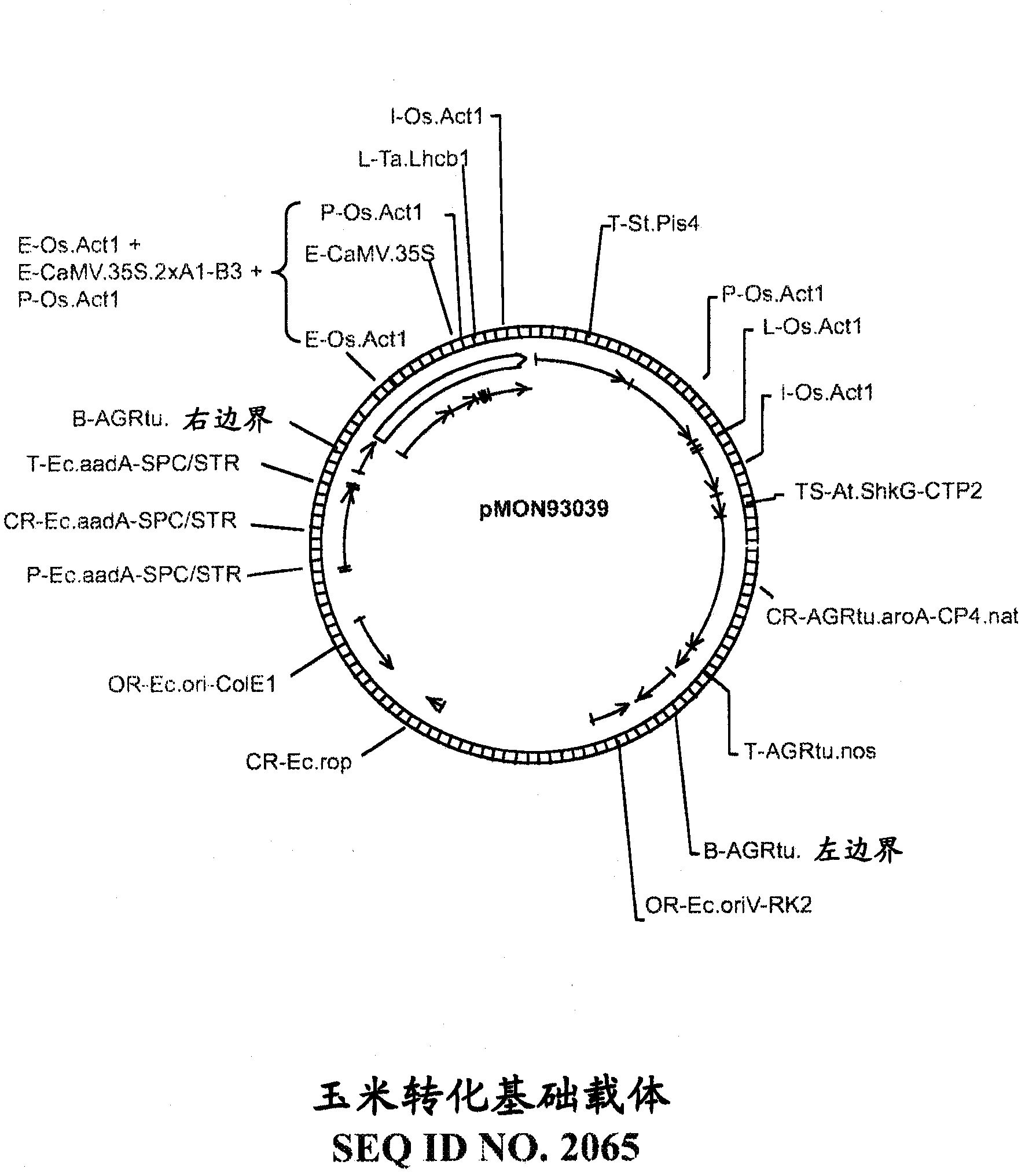
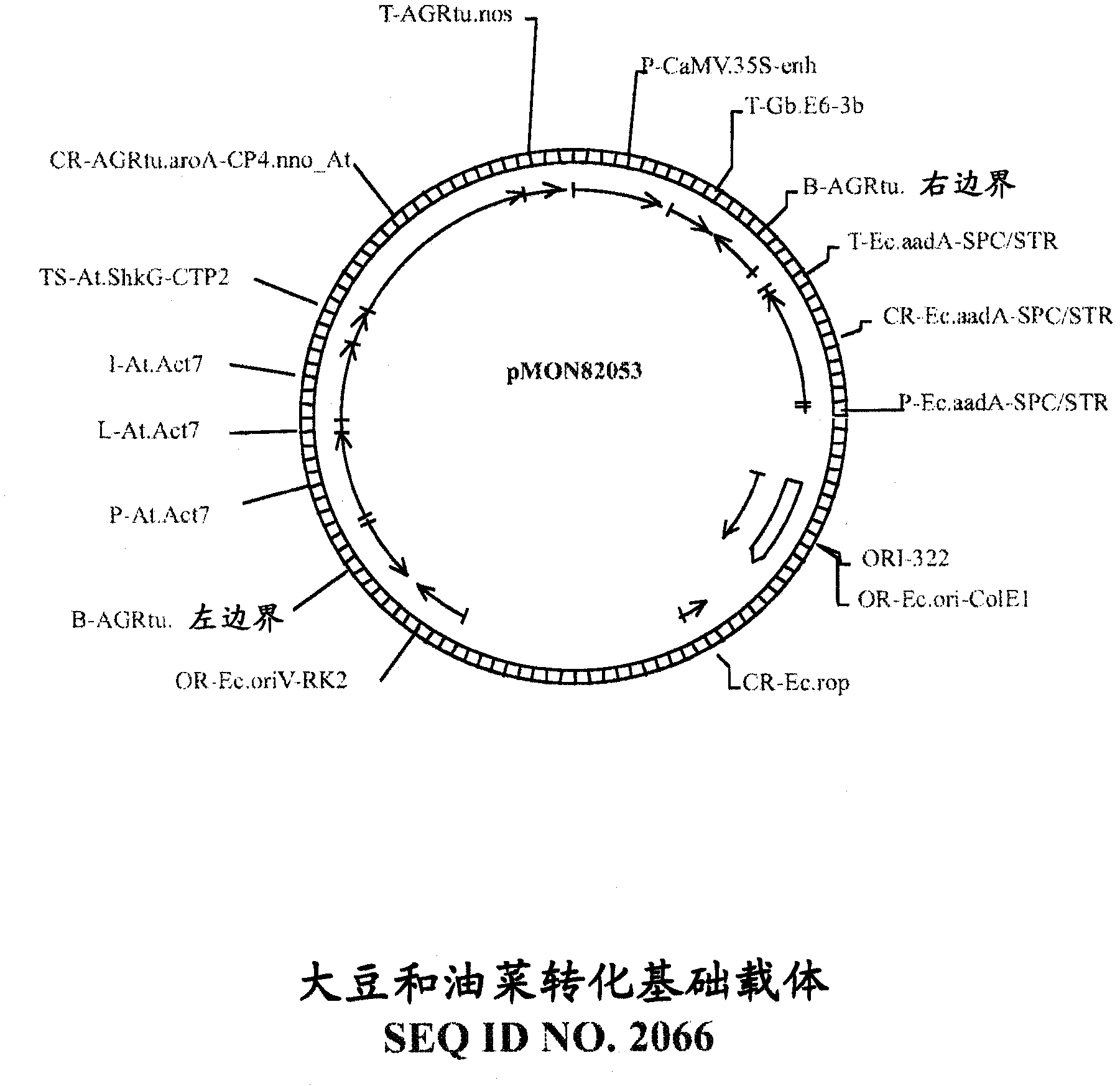
This invention provides recombinant DNA constructs and methods for manipulating expression of a target gene that is regulated by a small RNA, by interfering with the binding of the small RNA to its target gene. More specifically, this invention discloses recombinant DNA constructs encoding cleavage blockers, 5-modified cleavage blockers, and translational inhibitors useful for modulating expression of a target gene and methods for their use. Further disclosed are miRNA targets useful for designing recombinant DNA constructs including miRNA-unresponsive transgenes, miRNA decoys, cleavage blockers, 5-modified cleavage blockers, and translational inhibitors, as well as methods for their use, and transgenic eukaryotic cells and organisms containing such constructs.
Owner:MONSANTO TECH LLC
Reference internal type dual-luciferase reporter vector and application thereof
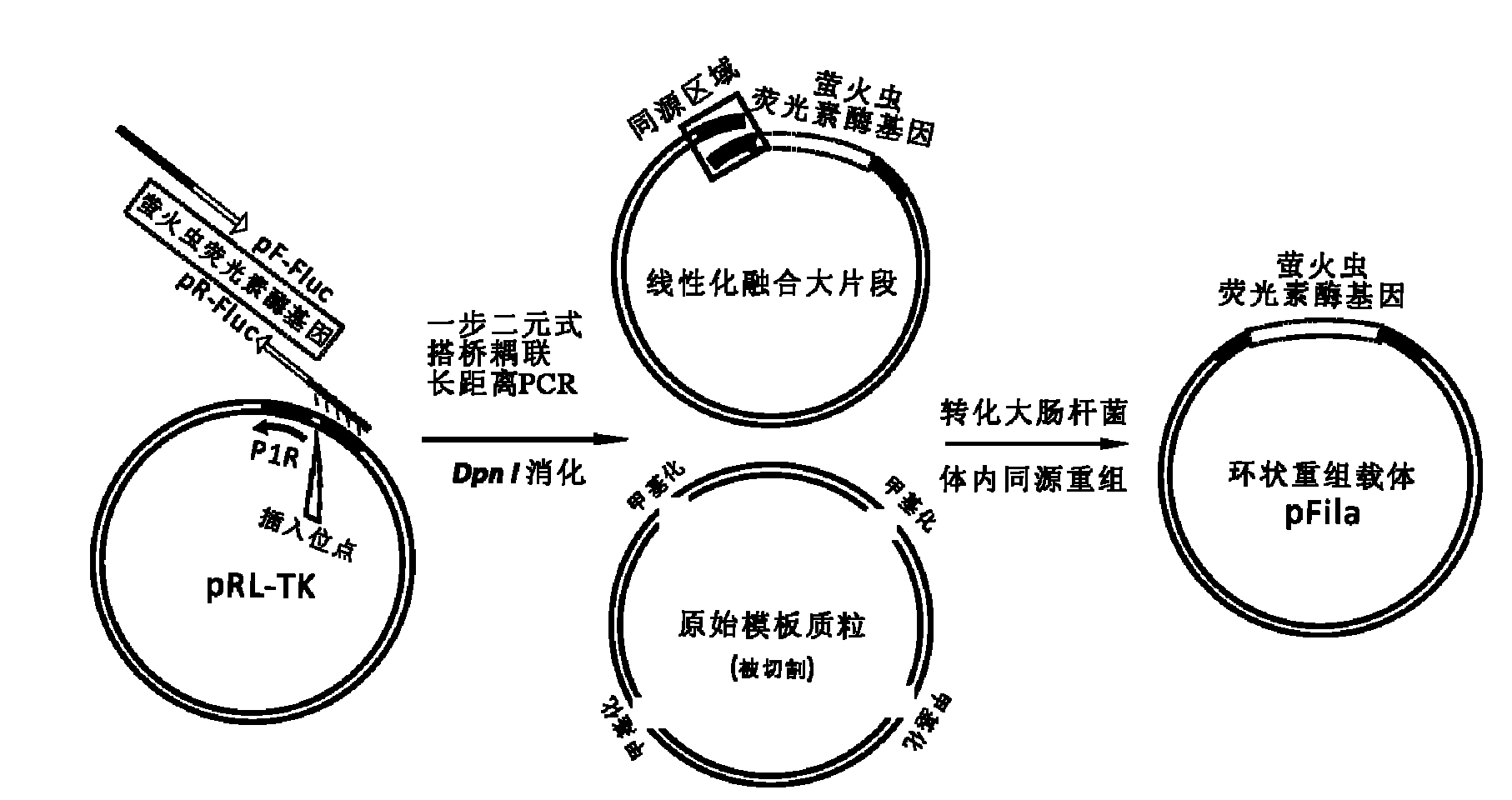
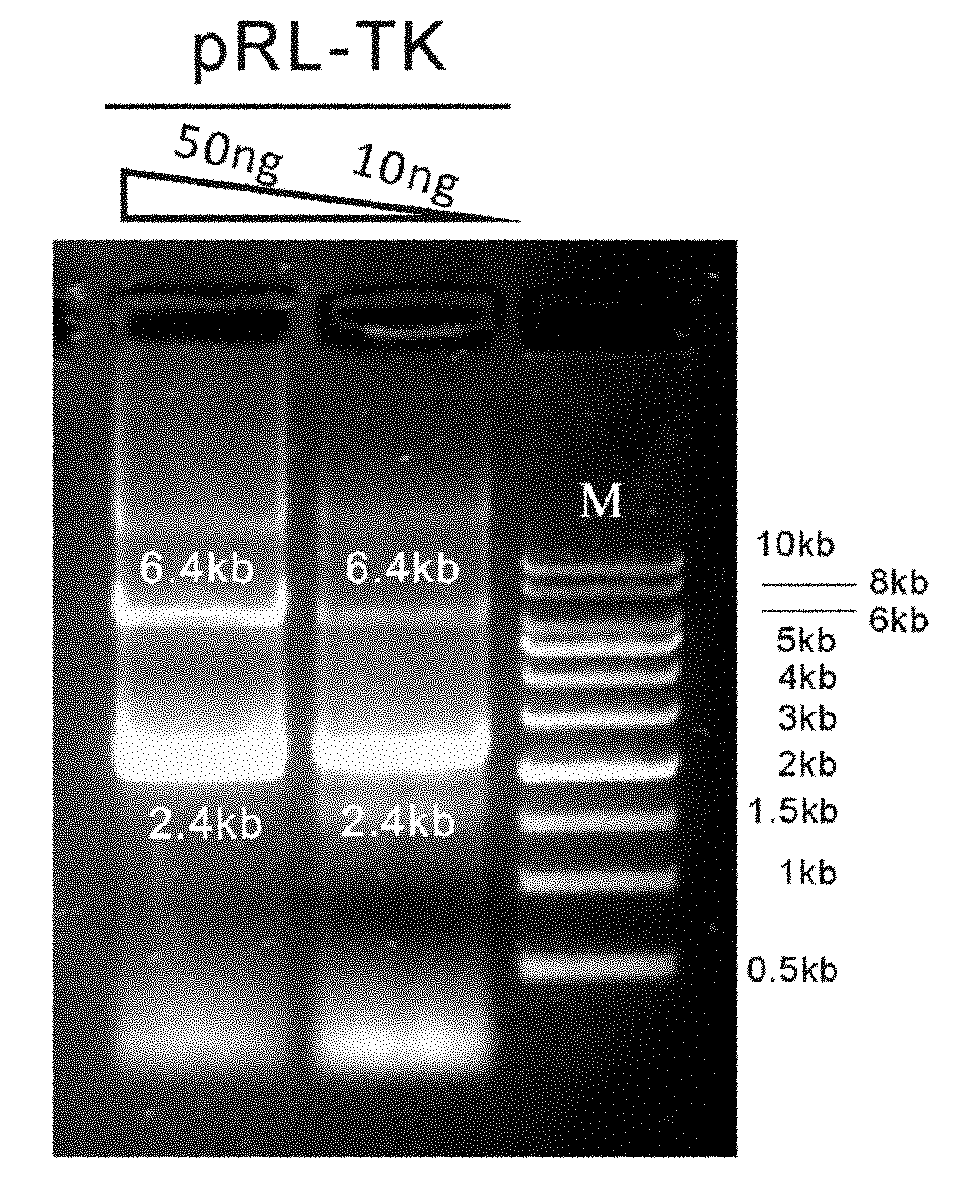
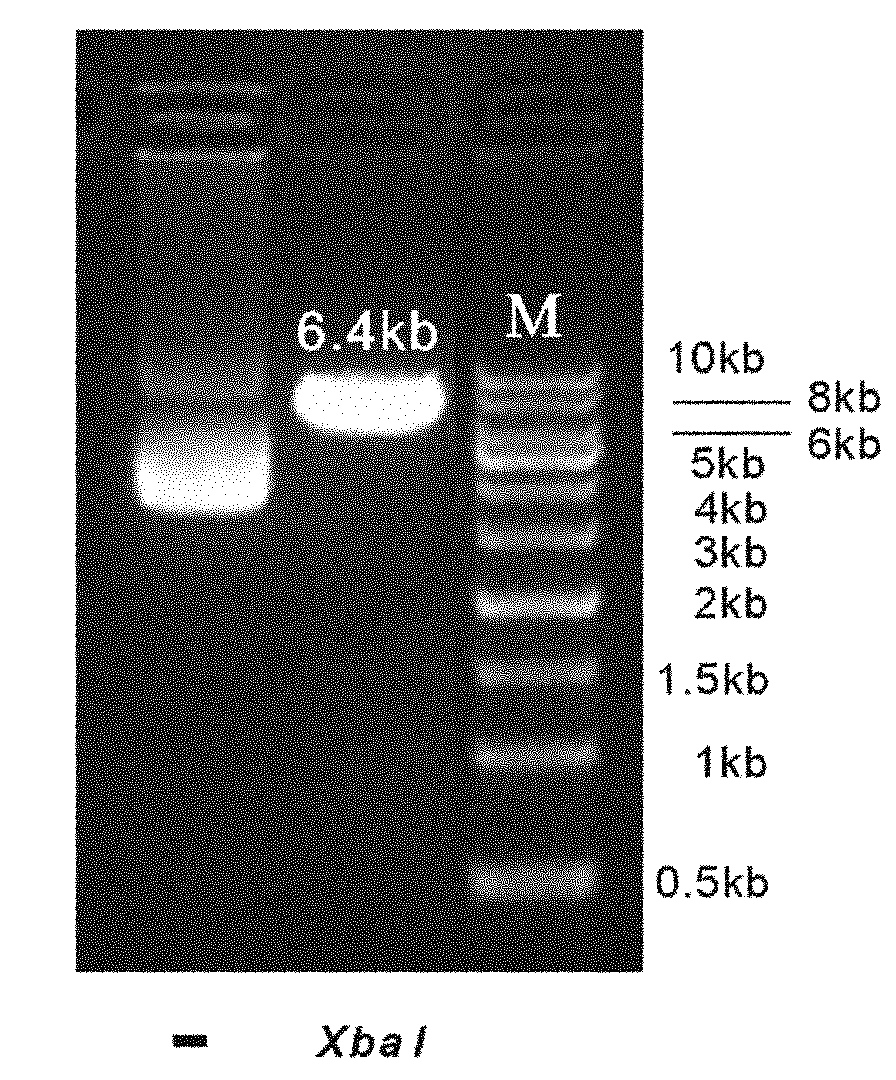
InactiveCN102094037AWill not interfere with each otherSimplify the transfection stepMicrobiological testing/measurementVector-based foreign material introductionMirna targetPolymerase chain reaction
The invention discloses a reference internal type dual-luciferase reporter vector and an application thereof. Firefly luciferase gene is fused between the renilla luciferase gene of the pRL-TK vector and the ampicillin resistance gene, the two luciferases are on the same vector, and the firefly luciferase gene is used as reference. The one-step binary bridging coupling long-distance polymerase chain reaction (PCR) and the Escherichia coli vivo homologous recombination method are utilized to place the firefly luciferase gene and the renilla luciferase gene on the same vector; and monoclonal sites are introduced in the 3' untranslated region of the renilla luciferase gene for conveniently cloning the target segment, thus the reference internal type dual-luciferase reporter vector can be constructed. The vector of the invention has the advantages of high repeatability, little multiple-pore variation, convenient operation and precise quantification. The vector of the invention is suitablefor the screening, identification and confirmation of the miRNA target molecule and also suitable for the quantitative analysis of the activity change of miRNA in cellular level.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
Method for researching oryza sativa and pathogen interaction mode
ActiveCN104911261AQuick and effective screeningStrong disease resistanceComponent separationMicrobiological testing/measurementDiseaseTotal rna
The invention discloses a method for researching an oryza sativa and pathogen interaction mode, and belongs to the field of plant biotechnology. The method for researching the interaction mode of oryza sativa in response to pathogen infection through integrated application of a transtriptome, a proteome and a miRNA group comprises the concrete steps: A, respectively taking oryza sativa leaf blade total RNAs, Small RNAs and total proteins of a disease-resistant material and a susceptible material at different time points before and after pathogen inoculation; B, respectively carrying out differential-expression spectrum analysis, differential proteomics analysis and miRNA identification and target gene analysis; and C, integrating and associating differential genes, proteins and miRNA target genes identified by the three ways, to reveal disease-resistant and susceptible reaction paths activated or inhibited in the oryza sativa and pathogen interaction process, so as to relatively systematically illuminate an oryza sativa-magnaporthe oryzae interaction mechanism. The method can quickly and effectively screen candidate genes associated with oryza sativa disease resistance.
Owner:SOUTH CHINA AGRI UNIV
Nucleic Acids and Libraries
InactiveUS20120058917A1Provides readout at protein levelPorous dielectricsLibrary screeningPolyadenylationTest sequence
Owner:KINGS COLLEGE LONDON
Compositions and methods of gene silencing in plants
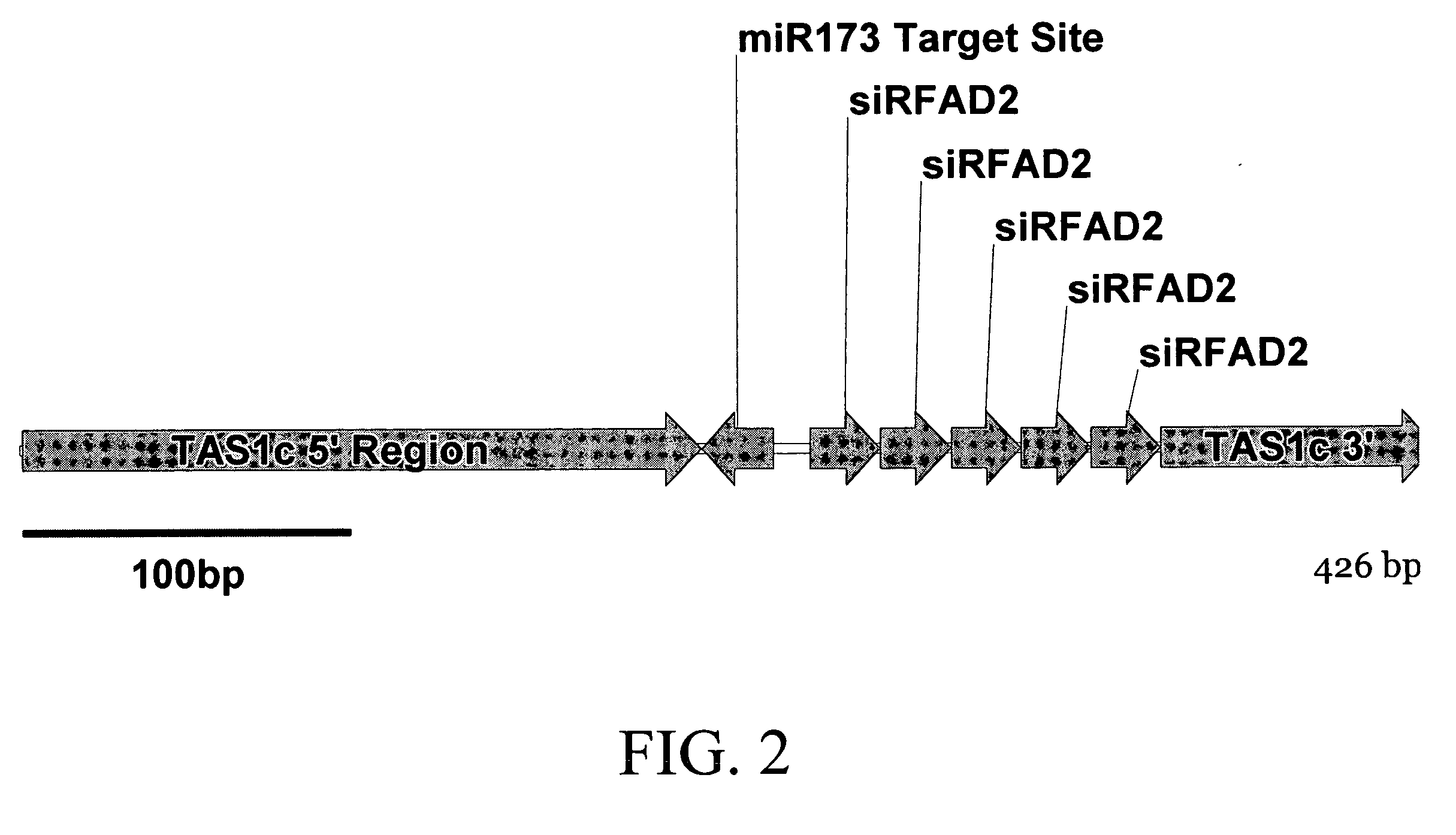
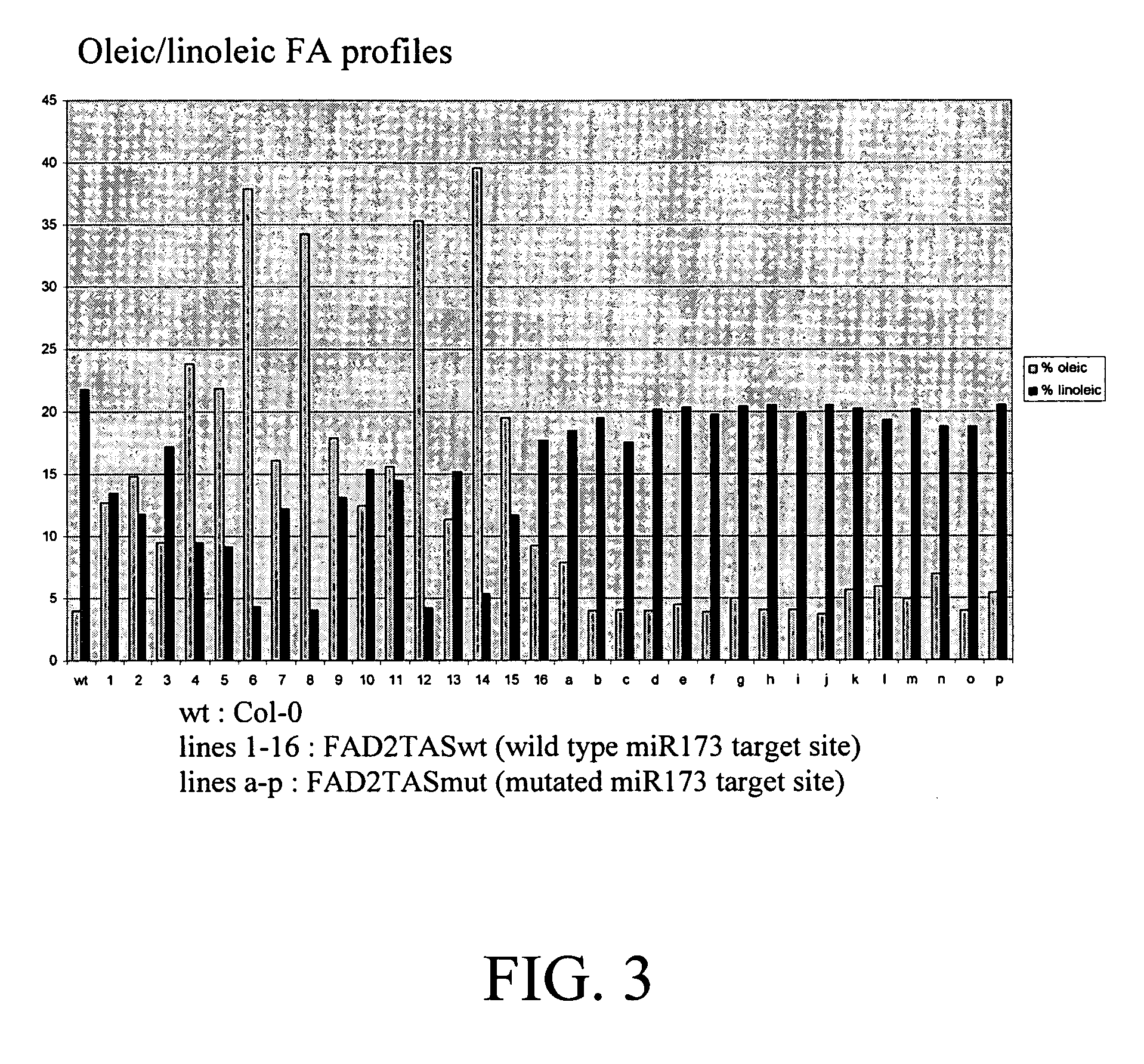
InactiveUS20150089688A1Reduce and inhibit expression of target genePrevention of stem breakOther foreign material introduction processesFermentationTrans-acting siRNAGene silencing
Compositions and methods for inducing gene silencing events in plants are disclosed. The compositions typical include a polynucleotide encoding an miRNA target sequence operably linked to a sequence of from a target gene, cDNA or mRNA, or fragment thereof. When expressed in the presence of an miRNA specific for the miRNA target sequence the compositions can induce production of trans-acting siRNA that silence the target of interest. Transgenic plants and preferred plant pathways that can be targeted using the disclosed methods and compositions are also disclosed.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS +1
Exogenous miRNA regulatory target gene prediction method containing three-dimensional free energy
ActiveCN108090327AImprove accuracyFeature input vector exactSequence analysisHybridisationFree energiesBinding site
The invention discloses an exogenous miRNA regulatory target gene prediction method containing three-dimensional free energy. In order to improve traditional sequence matching characteristics, statistical characteristics of the three-dimensional energy in a seed area and statistical characteristics of a spatial distribution penalty function at binding sites are newly provided, the characteristicsof the seed area binding sites represent the specific matching information of the binding sites, so that a constructed feature input vector is more accurate and more practical, and therefore the accuracy of miRNA target spot prediction is improved.
Owner:JILIN UNIV
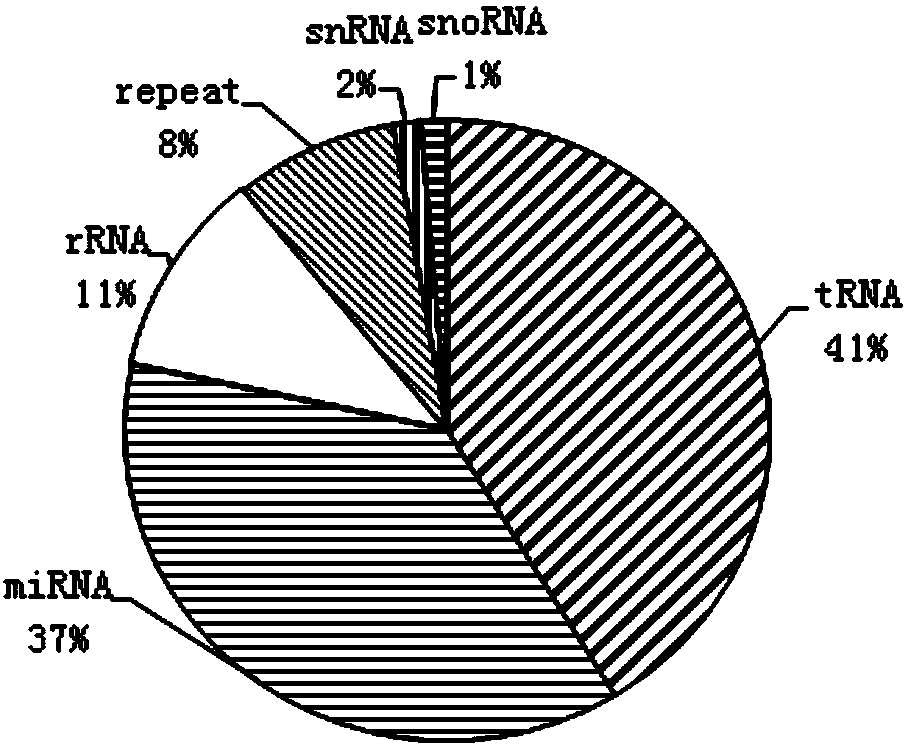
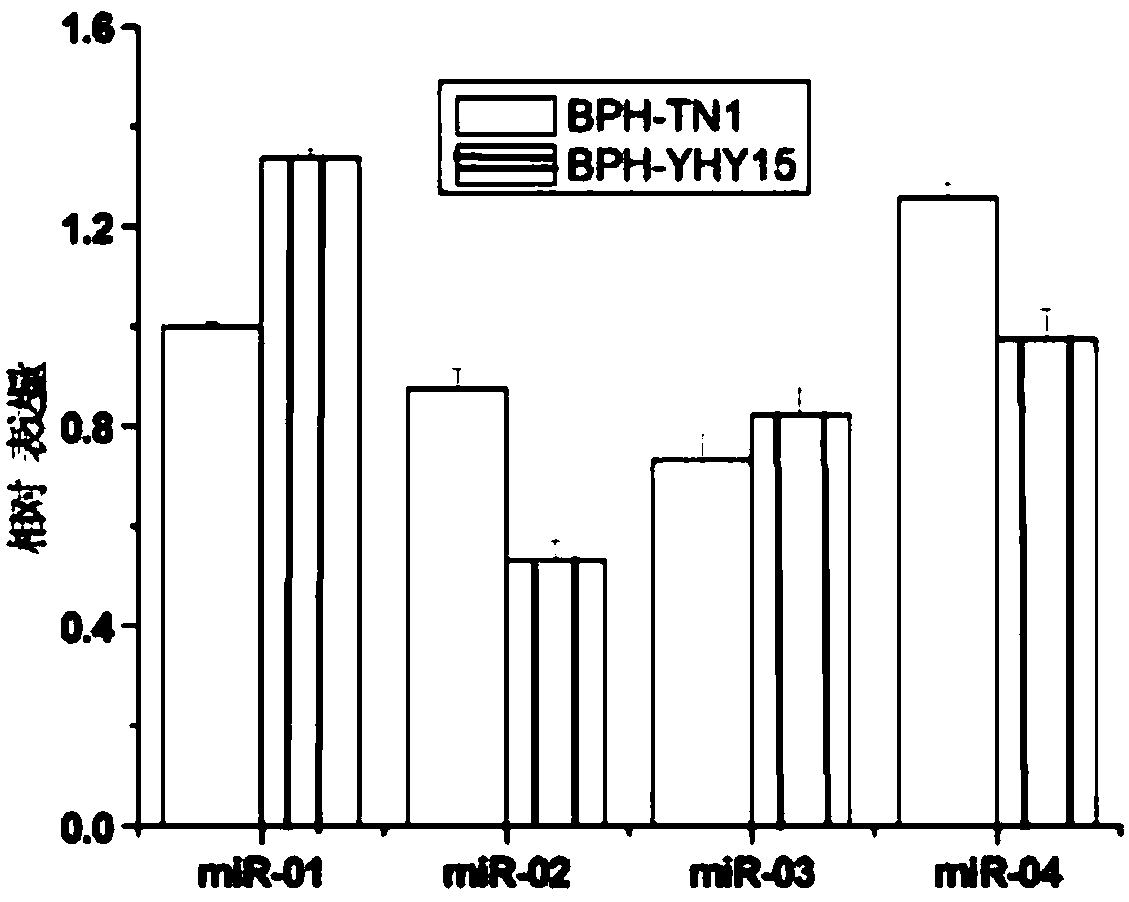
Method for screening MicroRNAs of nilaparvata lugens with effect on oryza sativa resistance adaptability
ActiveCN108504748AMicrobiological testing/measurementSpecial data processing applicationsBiologyMirna target gene
The invention discloses a method for screening MicroRNAs of nilaparvata lugens with effect on oryza sativa resistance adaptability, and relates to the bioengineering technology. The method comprises the following steps: (1) sample collection; (2) sequencing of small RNA; (3) pretreatment of sequencing raw data; (4) sequence alignment; (5) new miRNA prediction; (6) analysis of miRNA differential expression; (7) prediction of differential miRNA target genes; (8) synthesis of miRNA mimic, miRNA inhibitor and control chain; (9) microinjection and artificial feeding of miRNA mimic and miRNA inhibitor to nilaparvata lugens and verification of the function of miRNAs; (10) preliminary study on oryza sativa resistance adaptability change of the nilaparvata lugens after microinjection and artificialfeeding. Small RNAs of two nilaparvata lugens groups are deeply sequenced with a high-throughput sequencing technology, and are analyzed, identified and predicted according to subsequent bioinformatics, differences between the two nilaparvata lugens groups on insect-susceptible oryza sativa TN1 and insect-resistant oryza sativa YHY 15 are compared, the miRNA associated with host resistance adaptability is analyzed, screened and discovered, and the new method is provided for oryza sativa insect-resistant breeding.
Owner:INST OF FOOD CROPS HUBEI ACAD OF AGRI SCI
Pharmaceutical compositions comprising anti-miRNA antisense oligonucleotides
The invention provides pharmaceutical compositions comprising short single stranded oligonucleotides, of length of between 8 and 26 nucleobases which are complementary to human microRNAs selected from the group consisting of miR19b, miR21, miR122a, miR155 and miR375. The short oligonucleotides are particularly effective at alleviating miRNA repression in vivo. It is found that the incorporation of high affinity nucleotide analogues into the oligonucleotides results in highly effective anti-microRNA molecules which appear to function via the formation of almost irreversible duplexes with the miRNA target, rather than RNA cleavage based mechanisms, such as mechanisms associated with RNaseH or RISC.
Owner:SANTARIS PHARMA AS
Targets for human micro rnas in avian influenza virus (H5N1) genome
The present invention relates to targets for Human microRNAs in Avian Influenza Virus (H5N1) Genome and provides specific miRNA targets against H5N1 virus. Existing therapies for Avian flu are of limited use primarily due to genetic re-assortment of the viral genome, generating novel proteins, and thus escaping immune response. In animal models, baculovirus-derived recombinant H5 vaccines were immunogenic and protective, but results in humans were disappointing even when using high doses. Currently, two classes of drugs are available with antiviral activity against influenza viruses: inhibitors of the M2 ion channel, amantadine and rimantadine, and inhibitors of neuraminidase, oseltamivir, and zanamivir. There is paucity of information regarding effectiveness of these drugs in H5N1 infection. These drugs are also well known to have side effects like neurotoxicity. Thus there exists a need to develop alternate therapy for targeting the Avian flu virus (H5N1). The present invention addresses this need in the field.
Owner:COUNCIL OF SCI & IND RES
Target gene capable of improving regeneration capacity of plants, regulation molecules and application of target gene
ActiveCN111235175AClimate change adaptationVector-based foreign material introductionBiotechnologyMirna target
The invention relates to a target gene capable of improving regeneration capacity of plants, regulation molecules and application of the target gene and discloses a new target HAM (hairy meristem). MiRNA targeted to HAM plays a role of promoting plant regeneration by inhibiting HAM. The new technology which has universality and is capable of increasing regeneration rate of the plants effectively and conveniently, a new way is provided for improved breeding of the plants, and good application prospects are achieved.
Owner:CAS CENT FOR EXCELLENCE IN MOLECULAR PLANT SCI
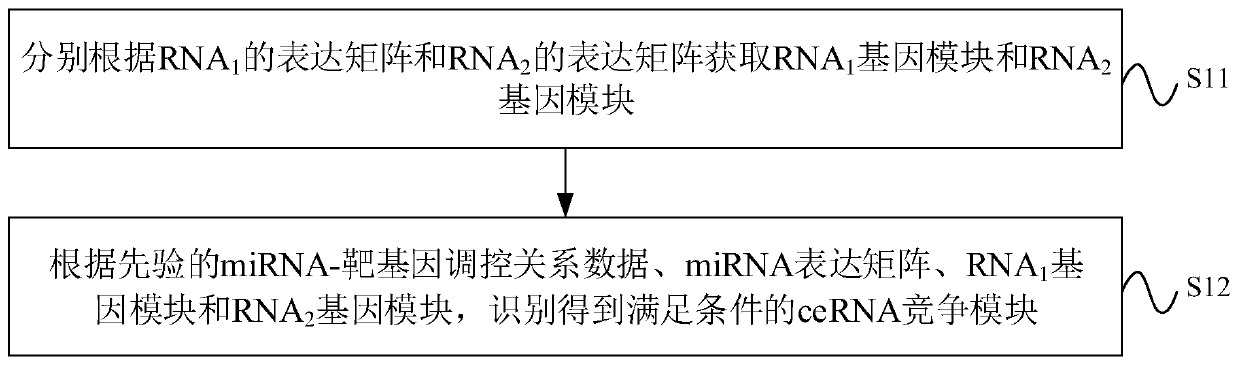
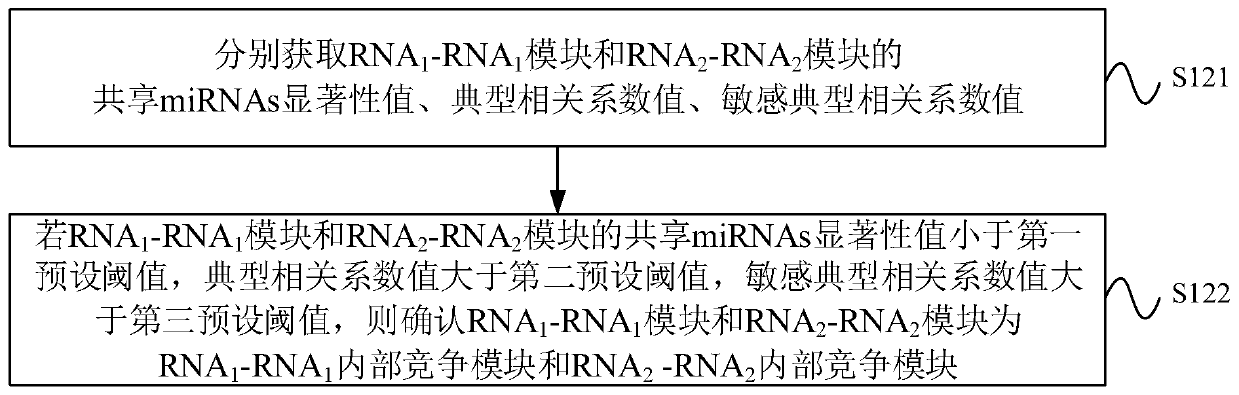
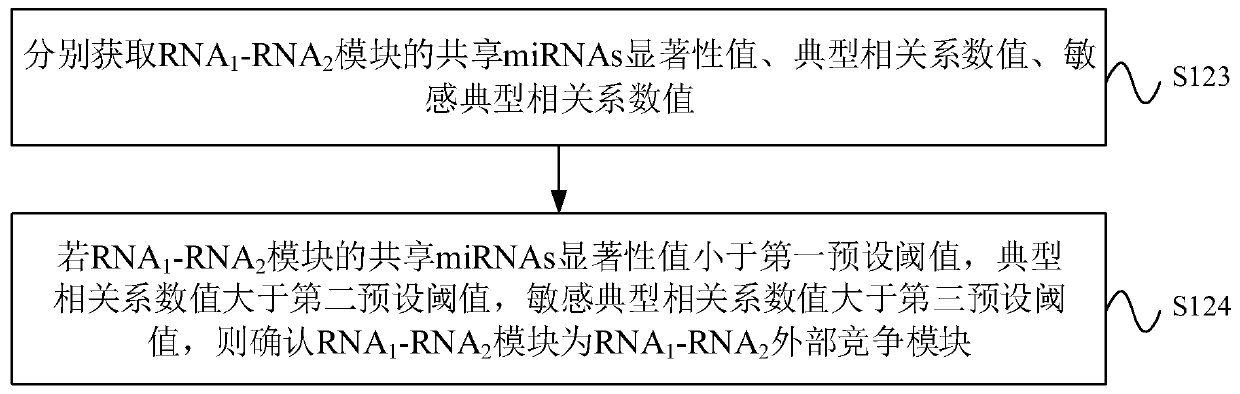
CeRNA competition model identification method and device, electronic equipment and storage medium
ActiveCN111383709AAccurate identificationCharacter and pattern recognitionProteomicsAlgorithmEngineering
The invention provides a ceRNA competition model identification method and device, electronic equipment and a storage medium, and relates to the field of gene identification. The ceRNA competition model identification method comprises the steps of obtaining an RNA1 gene module and an RNA2 gene module according to the expression matrix of the RNA1 and the expression matrix of the RNA2, wherein theRNA1 and the RNA2 are miRNA target genes; regulating relation data, a miRNA expression matrix, an RNA1 gene module and an RNA2 gene module according to the prior miRNA-target gene, and performing identification for obtaining the ceRNA competition module which satisfies a condition, wherein the ceRNA competition module comprises an inner competition module and an outer competition module. Because the inner and outer competition conditions of the ceRNA are considered, the inner and outer competition strength of the ceRNA and the influence of the miRNA to the inner and outer competition strengthsof the ceRNA are measured in a module level. Therefore, the method provided by the invention can accurately identify the ceRNA competition module.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA +1
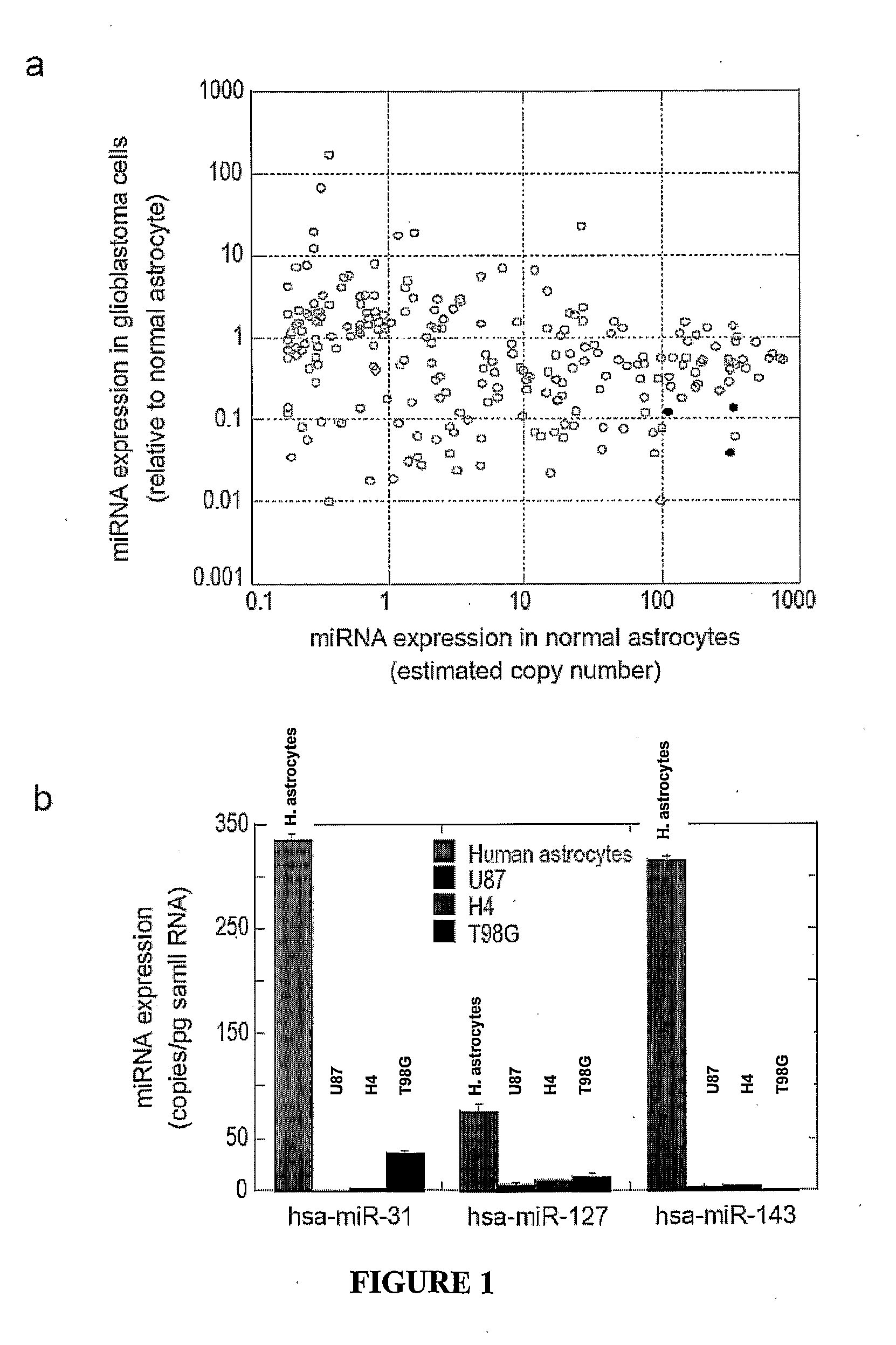
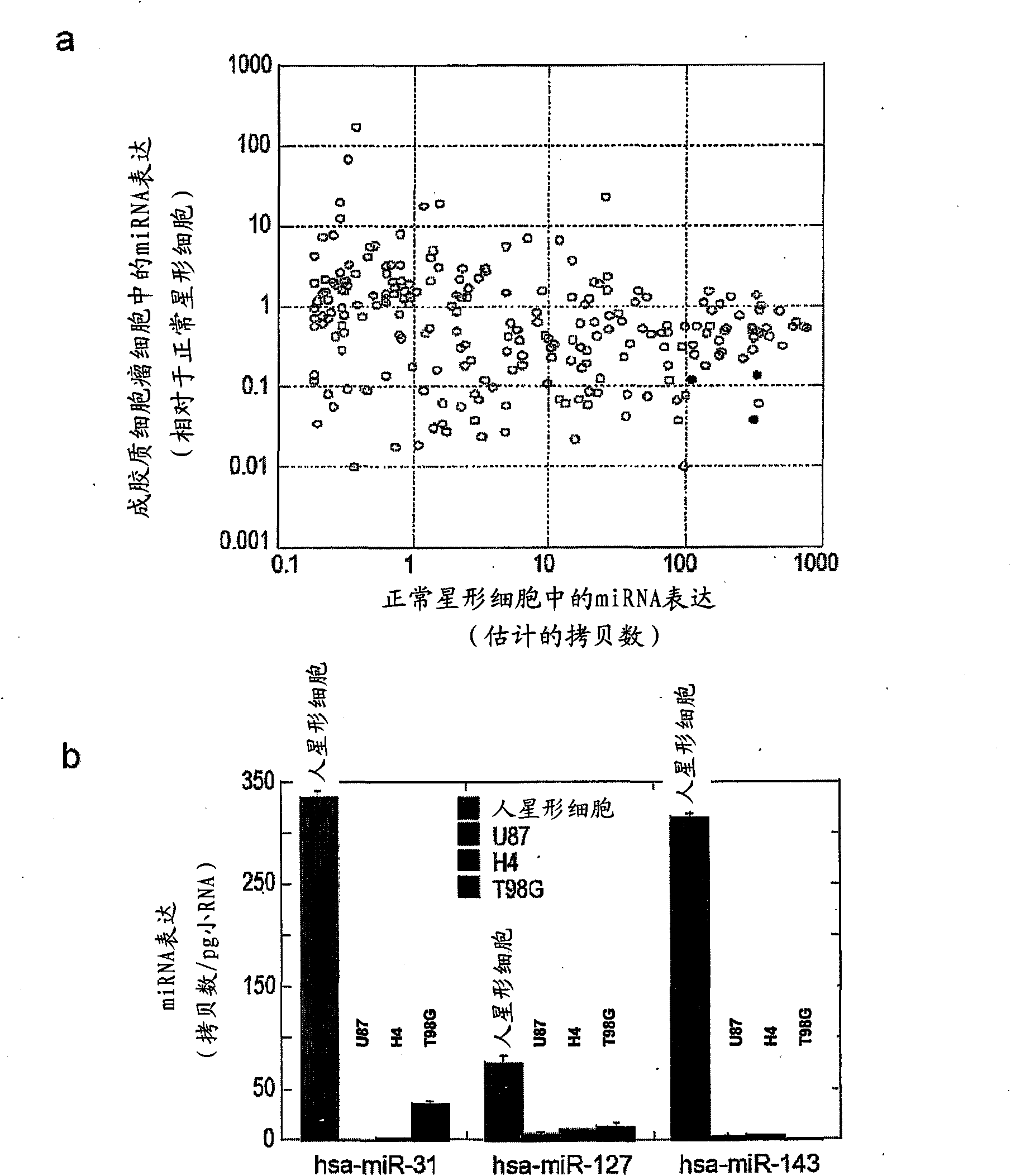
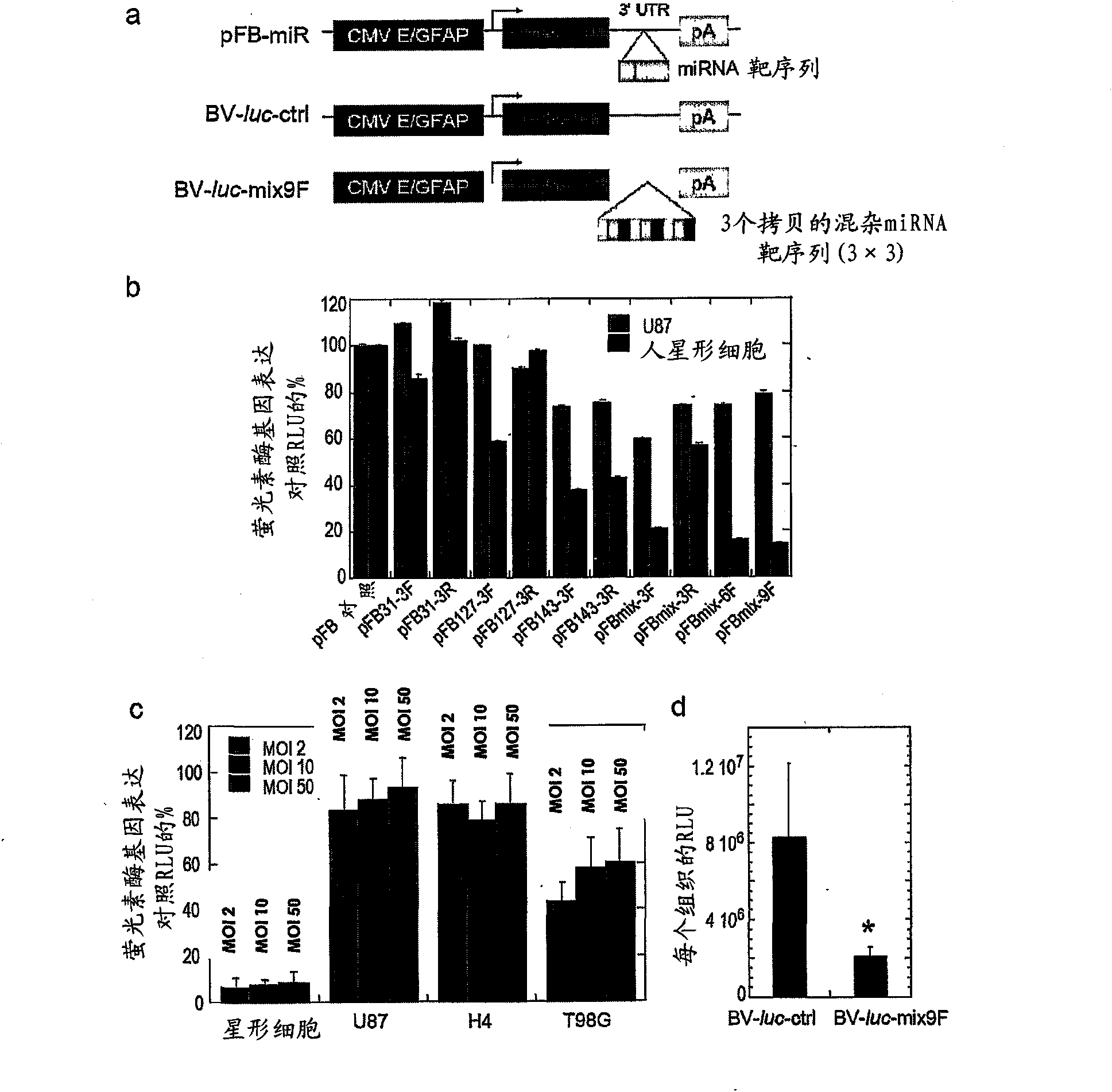
Nucleic acid molecule and method of targeting gene expression to gliomas
There is presently provided a nucleic acid molecule comprising a glial-specific promoter; a coding sequence for a transgene; and a plurality of miRNA target sites. Each miRNA target site binds an miRNA that is down-regulated in .a glioma cell compared to a normal glial cell, and the glial-specific promoter and the plurality of miRNA target sites are both operably linked to the coding sequence for the transgene.
Owner:AGENCY FOR SCI TECH & RES
Nucleic acid molecule and method of targeting gene expression to gliomas
Owner:AGENCY FOR SCI TECH & RES
Screening and identifying method of miRNAs (micro Ribonucleic Acids) of fetal fibroblasts of Saanen dairy goats
The invention discloses a screening and identifying method of miRNAs (micro Ribonucleic Acids) of fetal fibroblasts of Saanen dairy goats. The method comprises the following steps: obtaining the fetal fibroblasts of the Saanen dairy goats; extracting RNAs; constructing a library and carrying out high-throughput sequencing; and carrying out data processing. By adopting the high-throughput sequencing, 16395039total_reads are obtained, and redundant data is taken out to obtain clean_reads 16150181 containing Unique sRNAS 205857. Bioinformatic analysis is carried out to obtain 44 known miRNAs and 247 candidate new miRNAs. The quantity of miRNA target genes is 5401, and the quantity of sites of the target genes is 6069; and the quantity of candidate miRNA target genes is 8401, and the quantity of sites of the target genes is 10832.
Owner:TARIM UNIV
MicroRNA (micro ribonucleic acid) related to soybean phytophthora resistance
InactiveCN104726457AAchieve disease resistanceConvenient researchBiocideFungicidesRelation classificationResistant genes
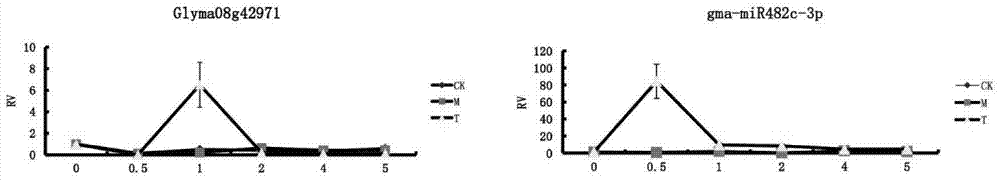
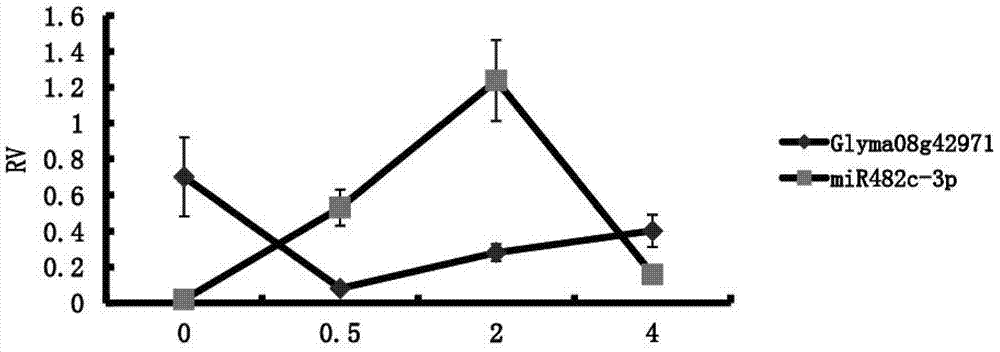
The invention discloses a microRNA (micro ribonucleic acid) related to soybean phytophthora resistance and belongs to the field of molecular biology of soybeans. 223 phytophthora-resistant genes and 256 miRNAs are obtained by means of screening known relevant disease-resistant immune genes. A phytophthora-resistance-related miRNA-Target predicting system is established by means of expression profile sequencing, rigorous bioinformatics analysis and real-time quantitative PCR (polymerase chain reaction) detection and can predict functions of the miRNAs, so that screening and identifying efficiency for soybean phytophthora resisting genes and the miRNAs is improved greatly. A MIRNA-target relation classification method is provided, that a Plant-pathogen interaction path plays a key role in phytophthora resisting is discovered, and that a gma-miR482c-3p serves as a main ingredient of the Plant-pathogen interaction path and is closely related to the soybean phytophthora resistance is verified.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
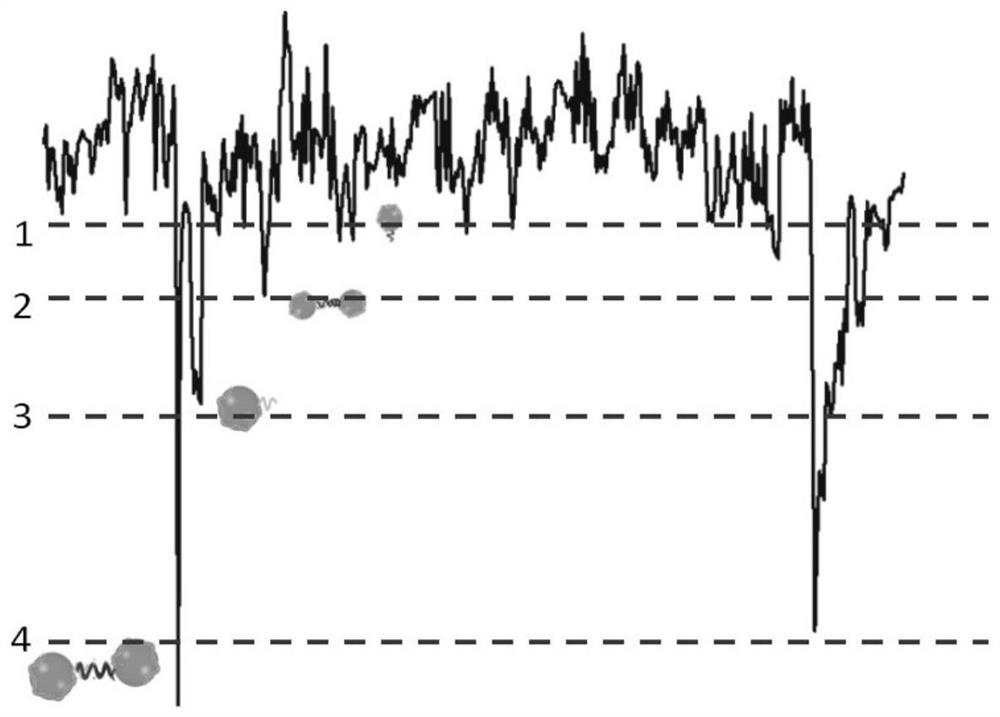
MiRNA detection method based on a solid nanopore sensor, detection probe and kit.
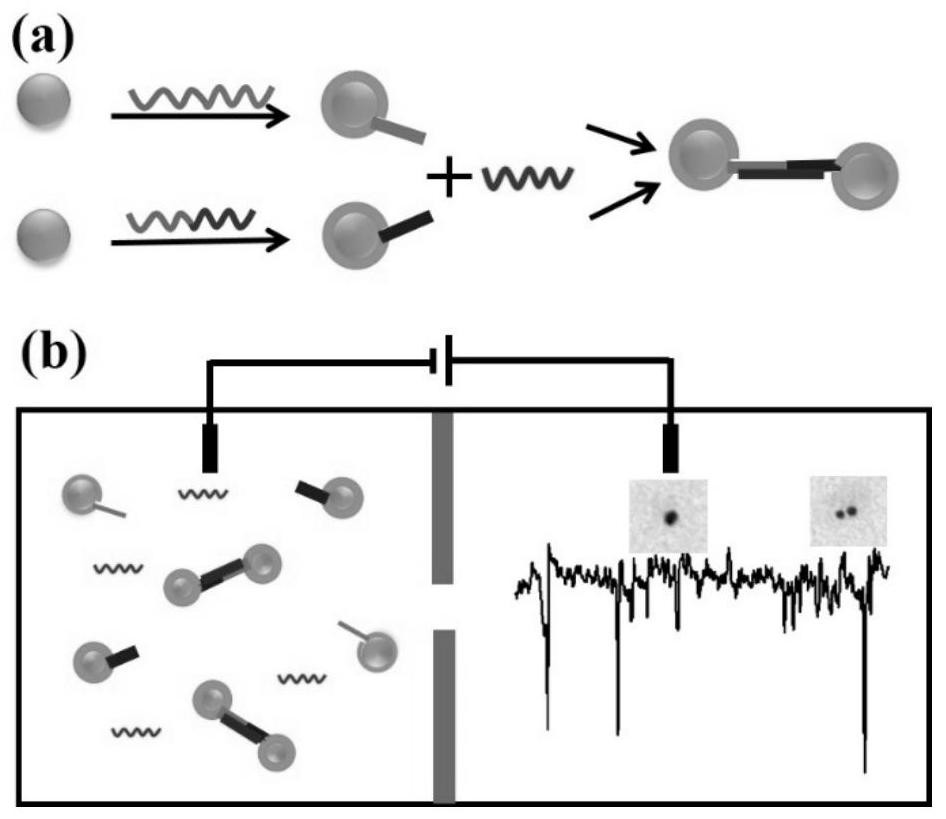
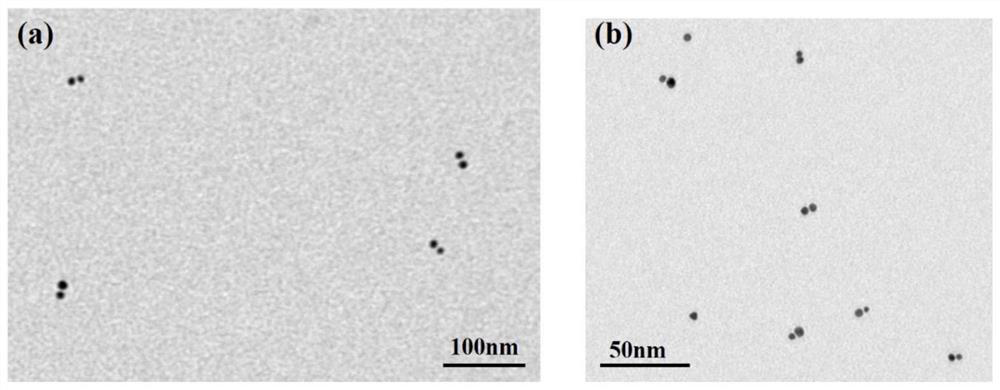
PendingCN112795628ARealize multiple detectionImprove early diagnosisMicrobiological testing/measurementDNA/RNA fragmentationBiological targetLung cancer
The invention discloses a miRNA detection method based on a solid nanopore sensor, a detection probe and a kit. The detection method provided by the invention comprises the following steps: taking gold nanoparticles as a carrier, and carrying out DNA probe monomolecular marking on different gold nanoparticles. When the probe and the target miRNA coexist, the DNA probe and the miRNA target molecule are hybridized and complemented, and the probe is induced to form a gold nanoparticle dimer. When the gold nanoparticle monomer and the gold nanoparticle dimer pass through the nanopore sensor, different current track characteristic signals are generated, so that the miRNA target molecule is identified. The method can be used for multi-element detection of multiple lung cancer marker miRNA biological target molecules at the same time. According to the invention, the problem that a solid-state nanopore sensor cannot detect miRNA small biomolecules is solved, multi-element joint detection of multiple lung cancer marker miRNA target molecules is realized, and the application of the silicon nitride solid-state nanopore in the field of single molecule detection is expanded.
Owner:NANJING UNIV OF POSTS & TELECOMM
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com
![Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks](https://images-eureka.patsnap.com/patent_img/2ceb1e3a-4328-4313-b6c3-95d3a50590fe/DEST_PATH_IMAGE001.png)
![Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks](https://images-eureka.patsnap.com/patent_img/2ceb1e3a-4328-4313-b6c3-95d3a50590fe/DEST_PATH_IMAGE002.png)
![Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks](https://images-eureka.patsnap.com/patent_img/2ceb1e3a-4328-4313-b6c3-95d3a50590fe/DEST_PATH_IMAGE003.png)