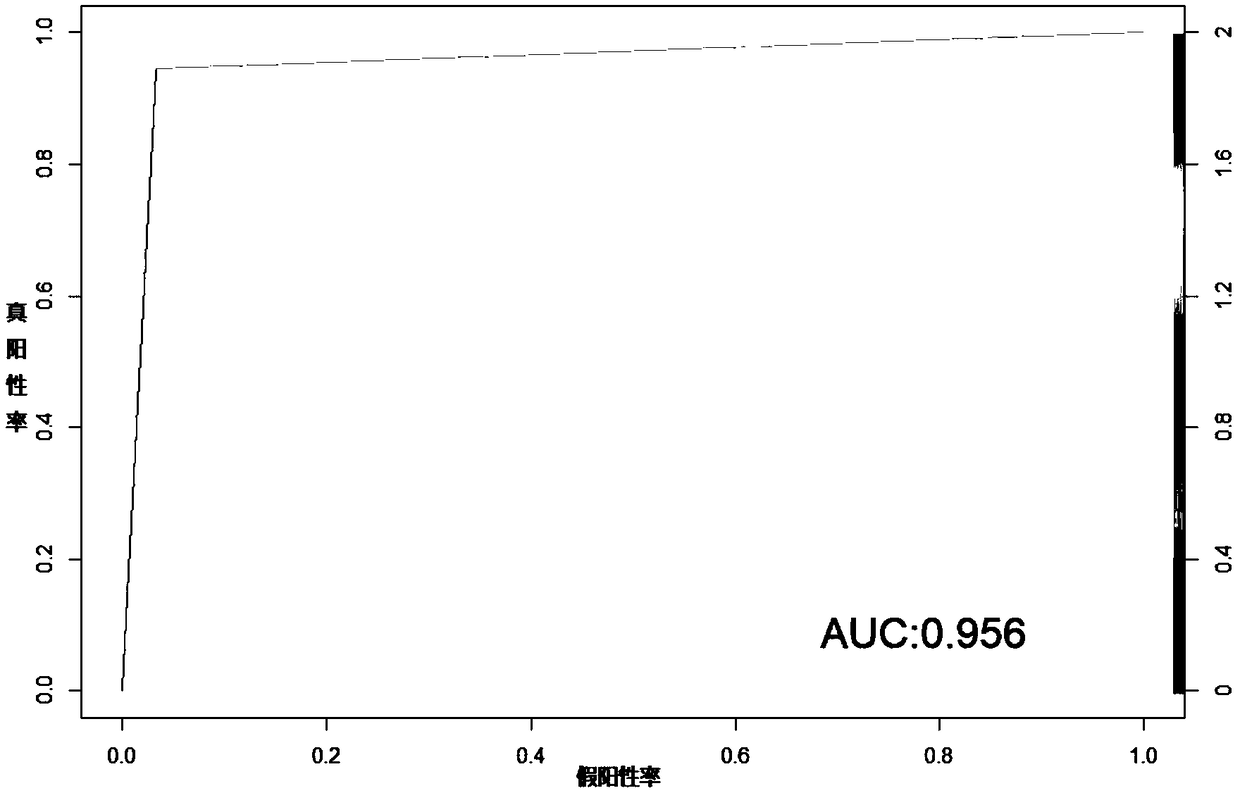
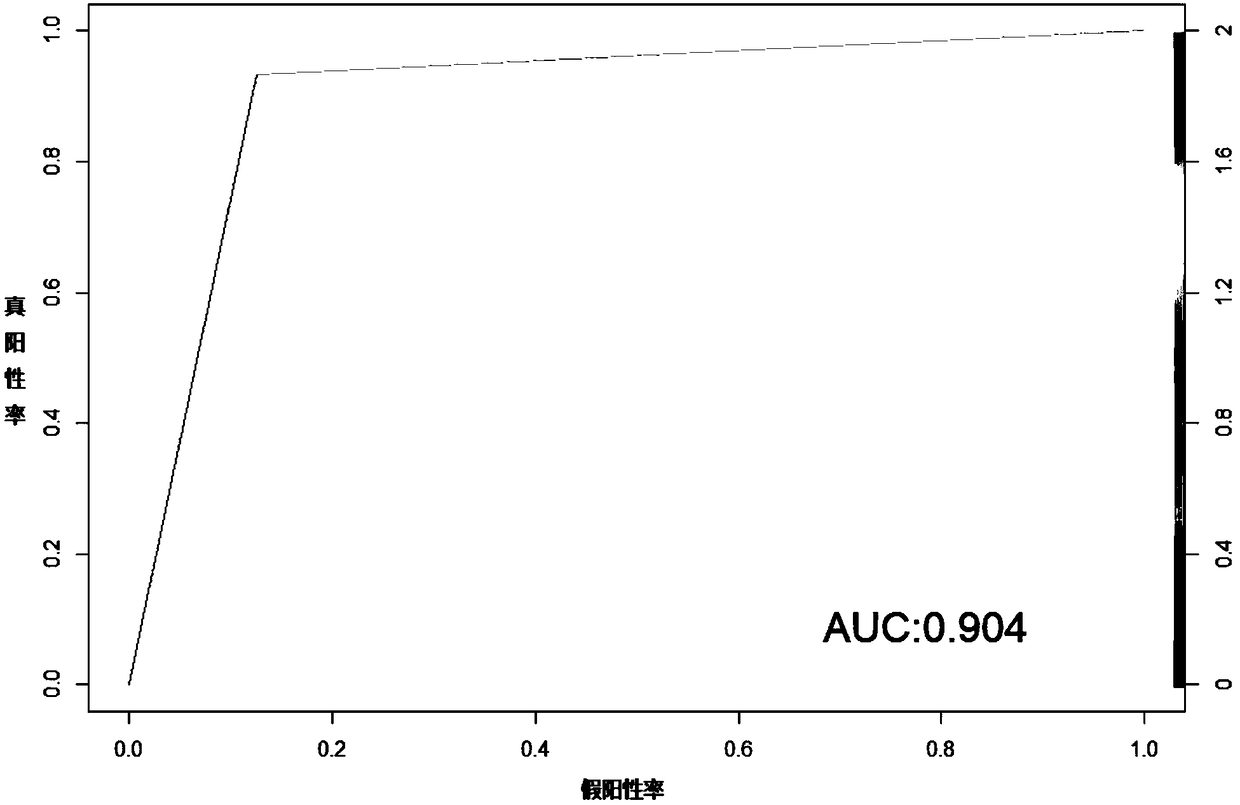
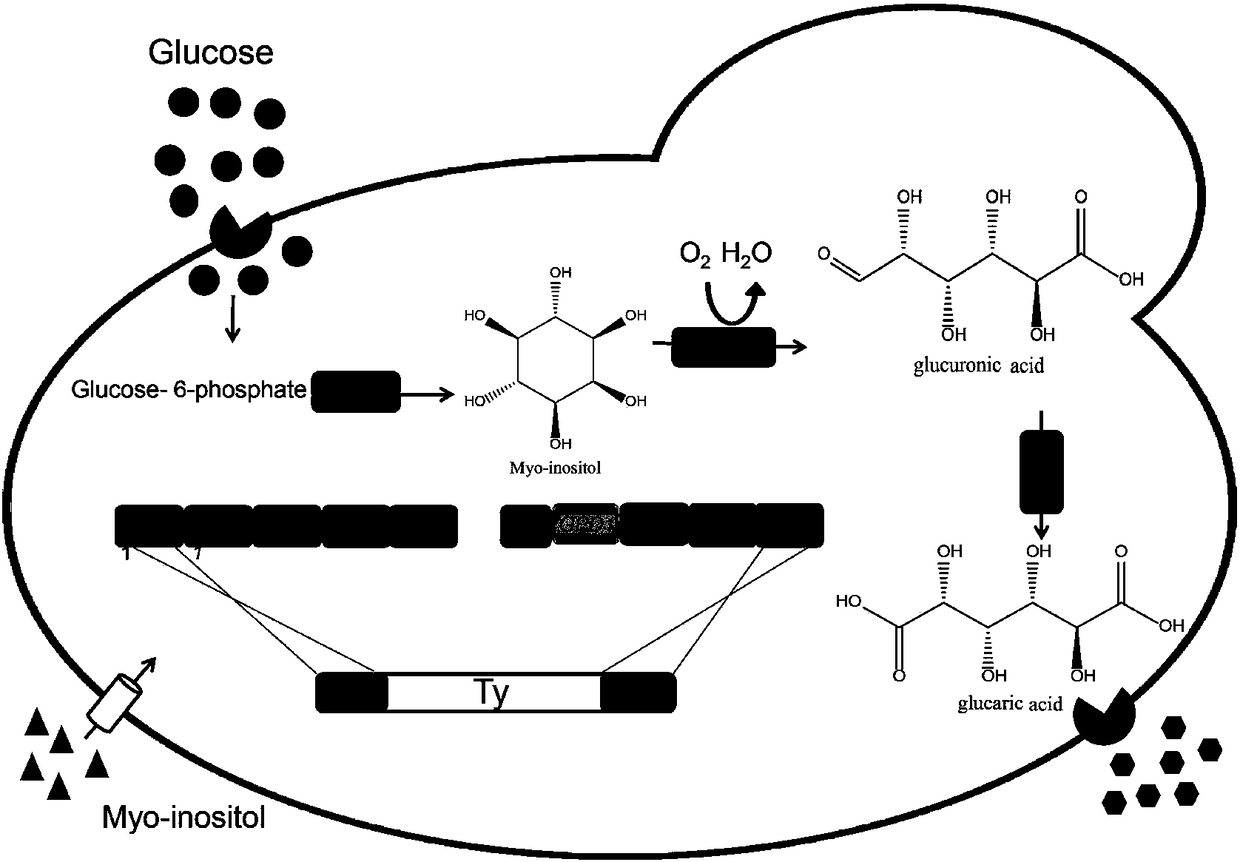
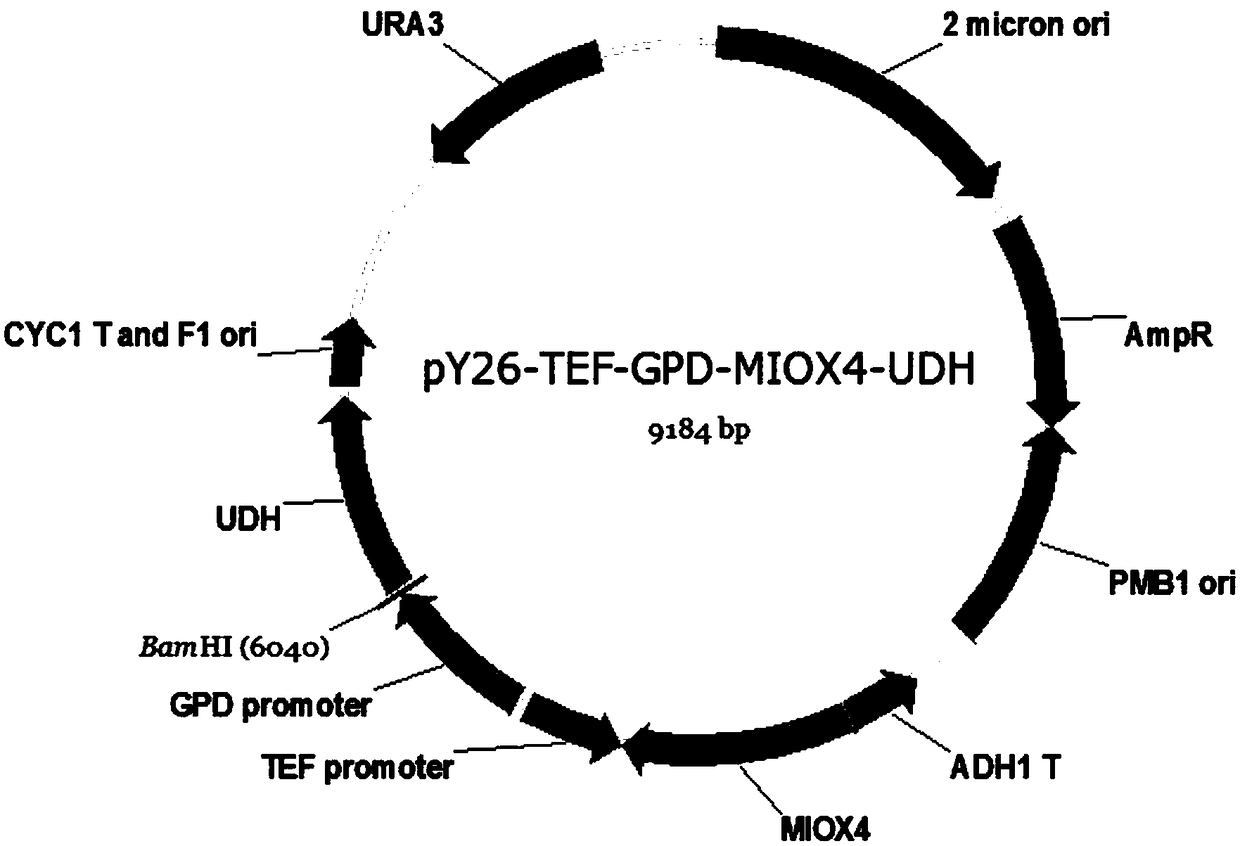
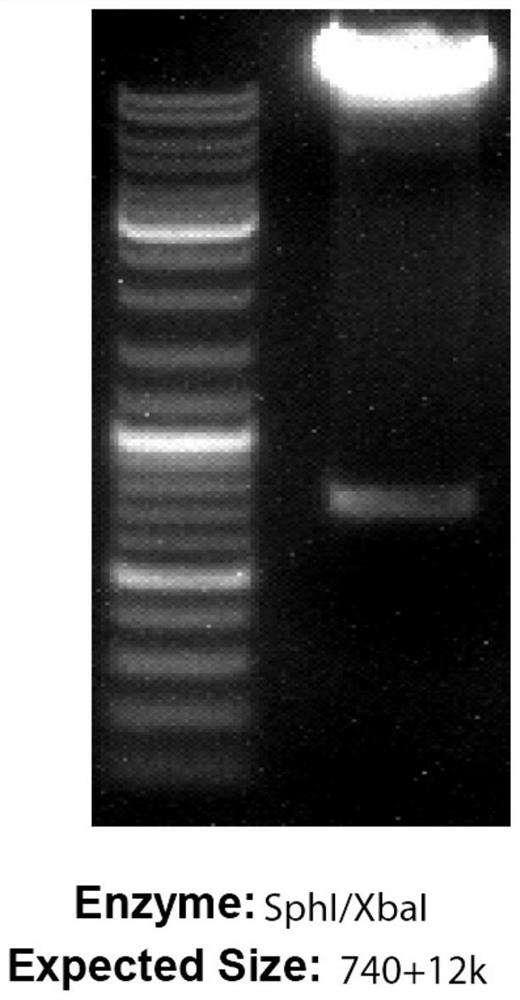
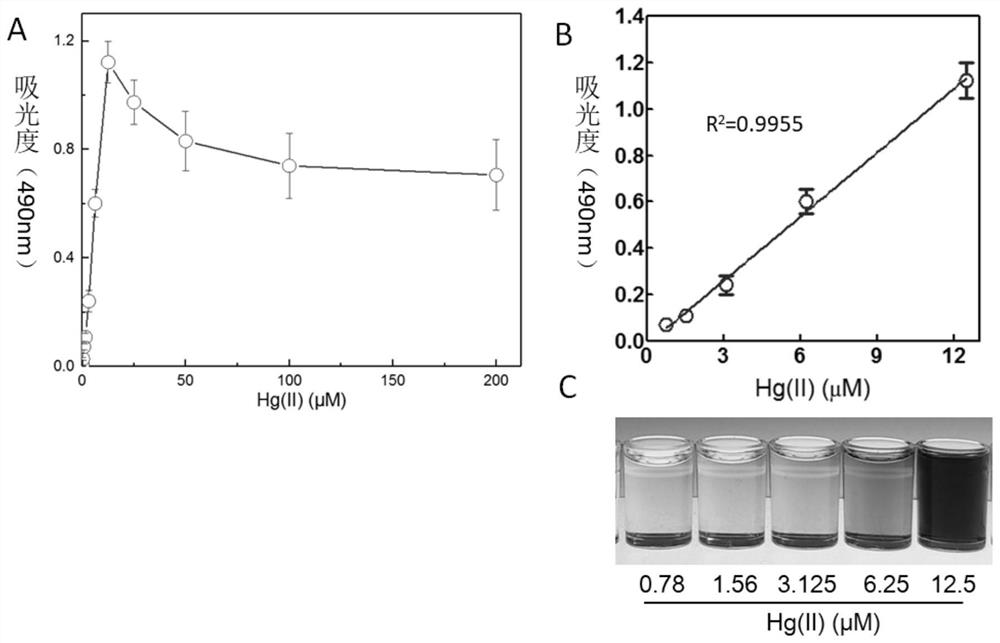
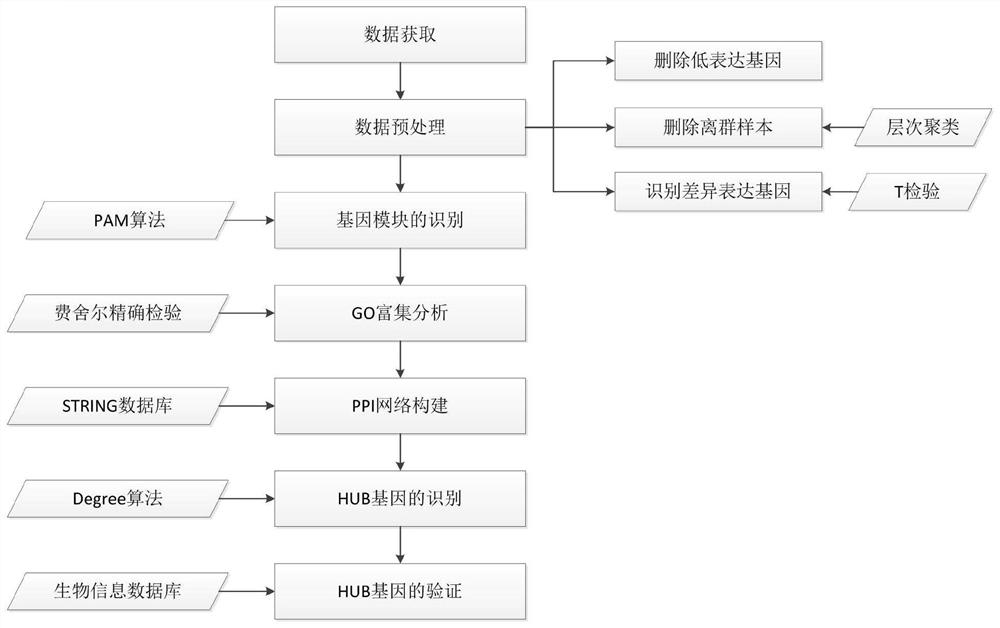
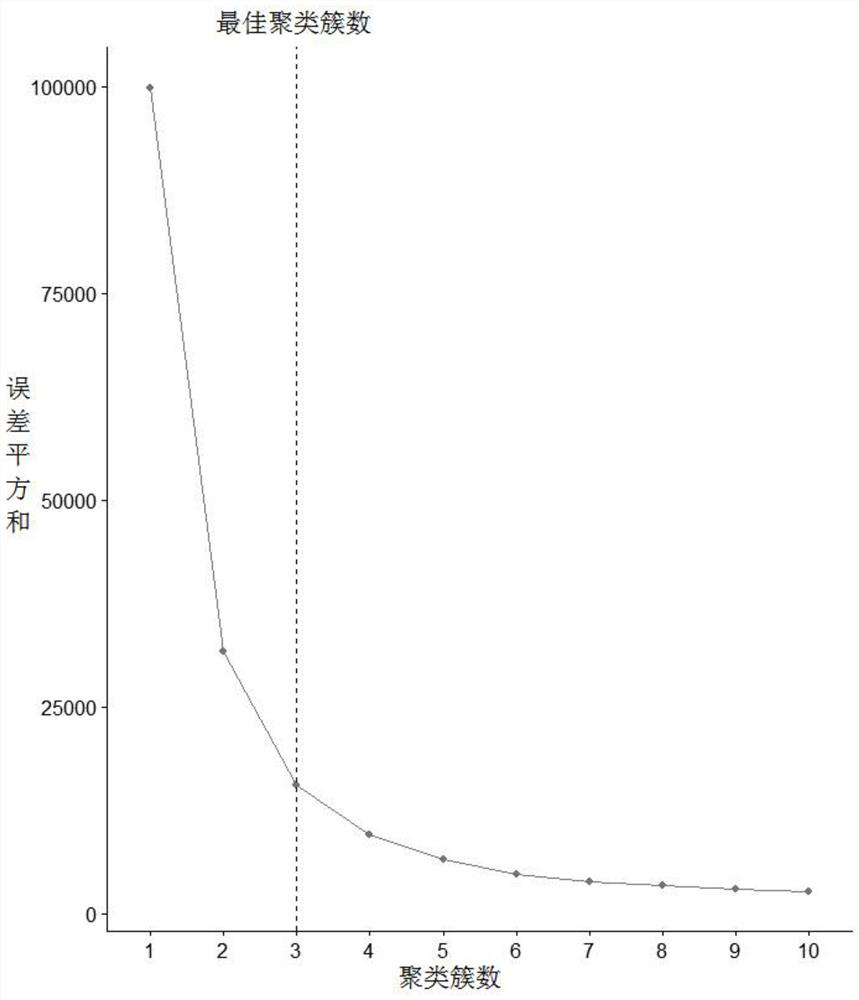
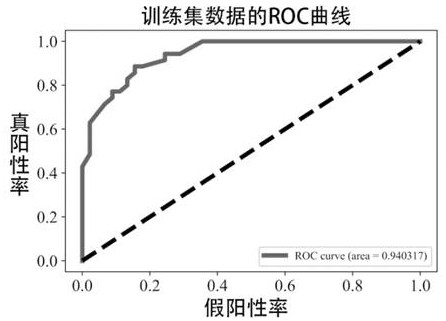
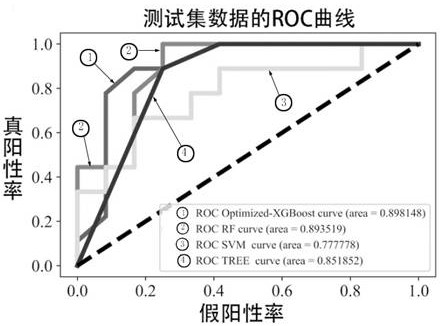
Patents
Literature
44 results about "Gene module" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The Human Gene module is an active collection of candidate genes identified through genetic association studies, genes linked to syndromic autism, and genes in which rare mutations have been found that are linked to autism.
Modulation of novel immune checkpoint targets
PendingUS20190255107A1Decrease in exhausted T cell phenotypeIncrease T cell activationAntibacterial agentsCell receptors/surface-antigens/surface-determinantsDiseaseClinical efficacy
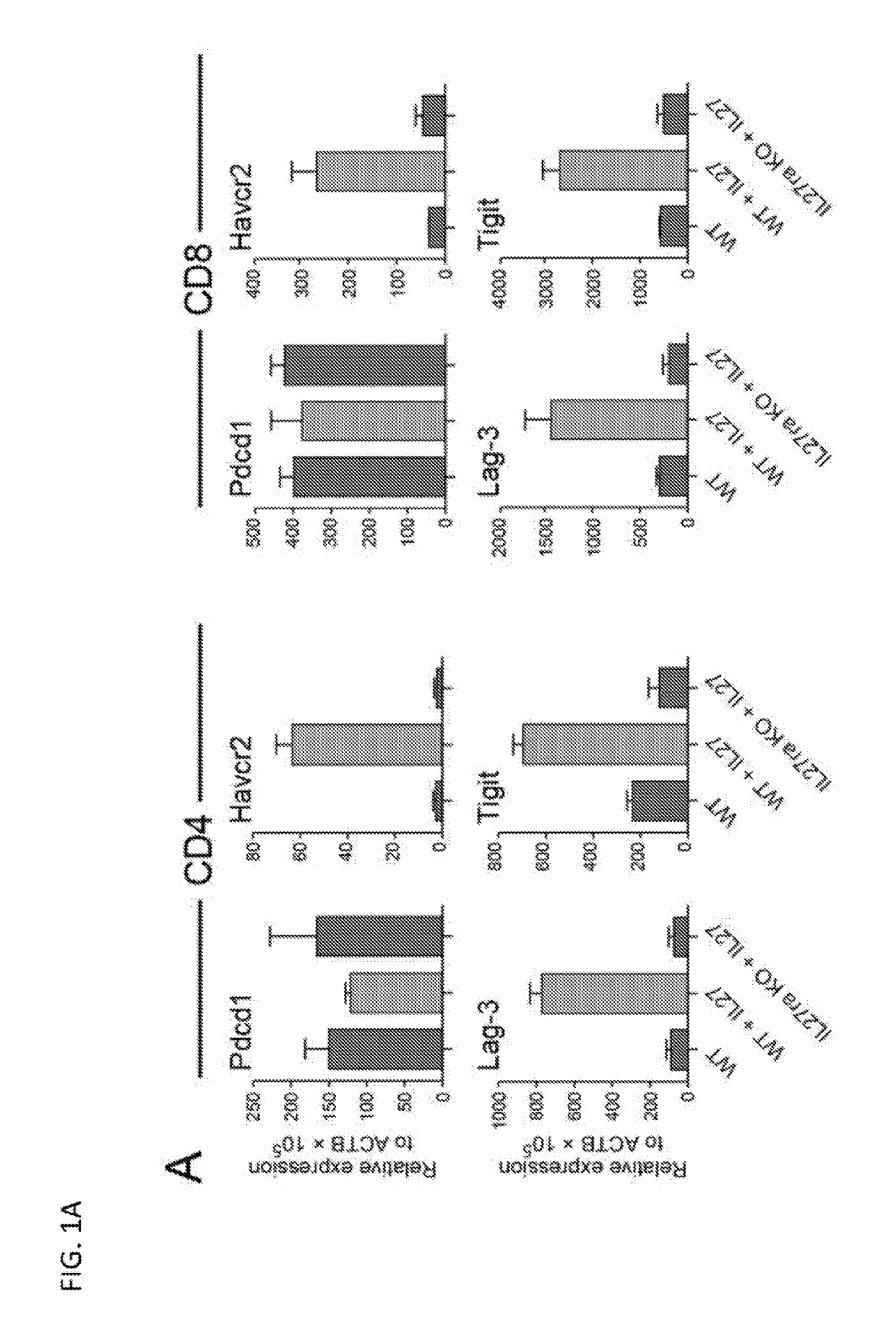
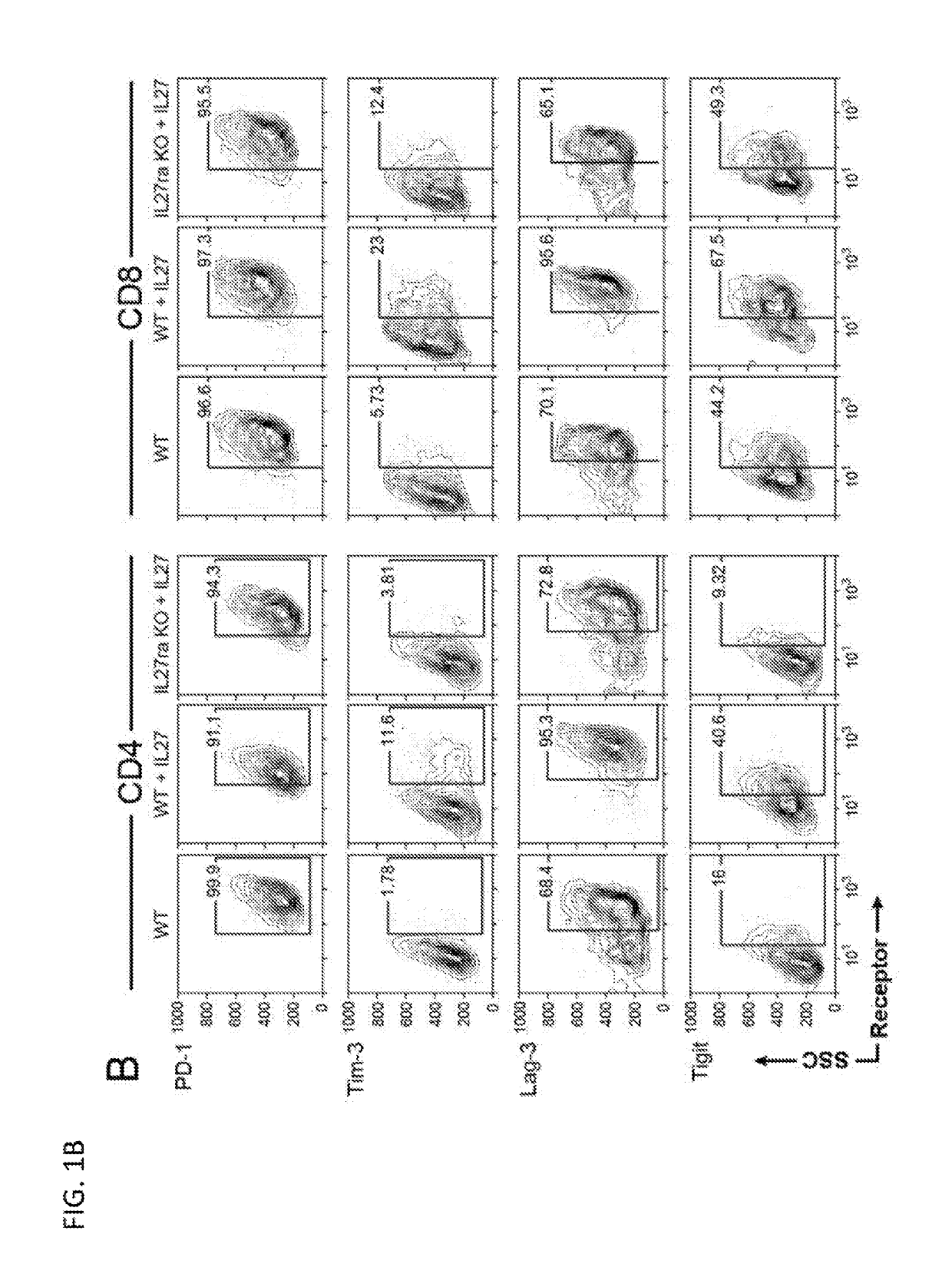
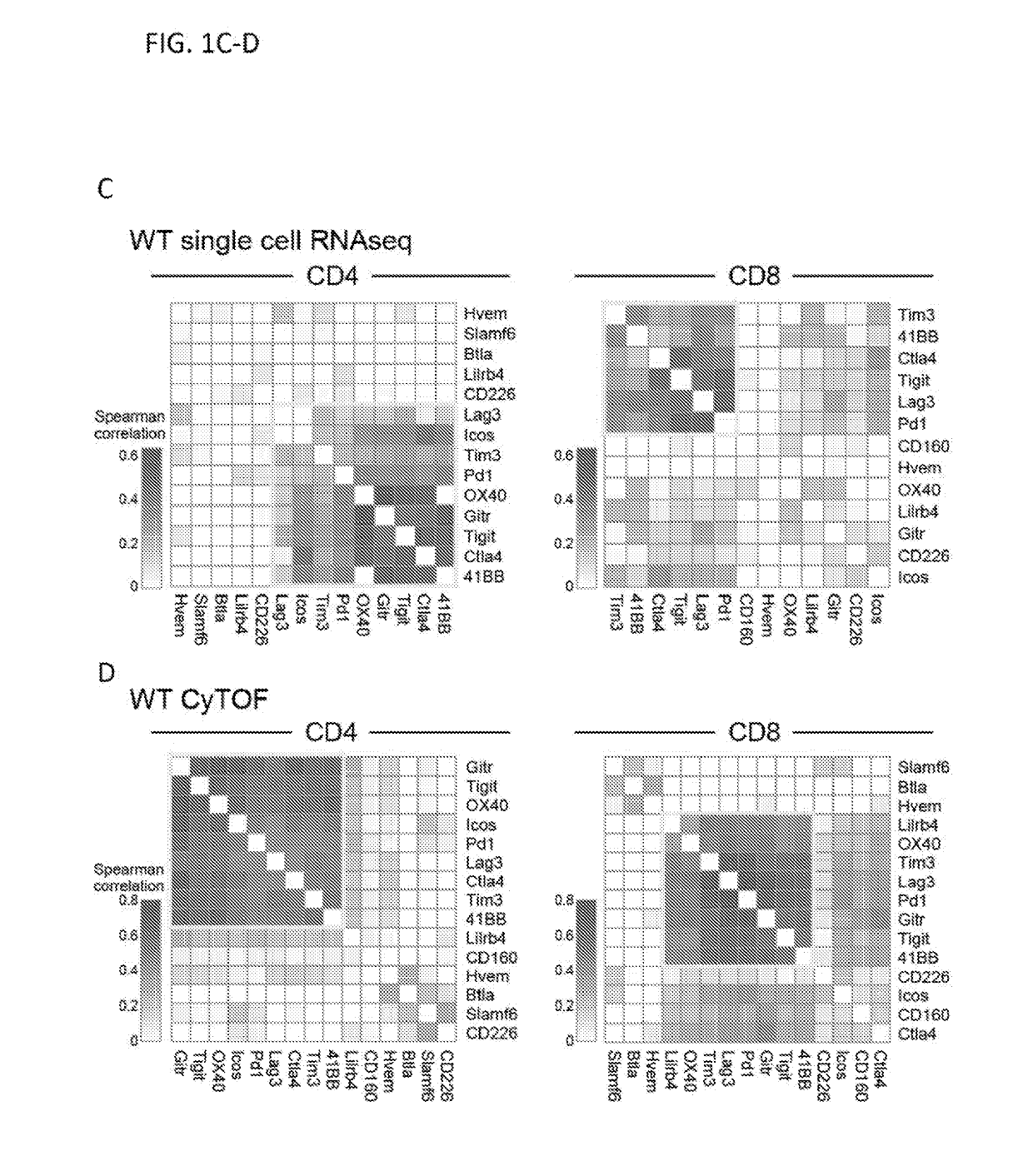
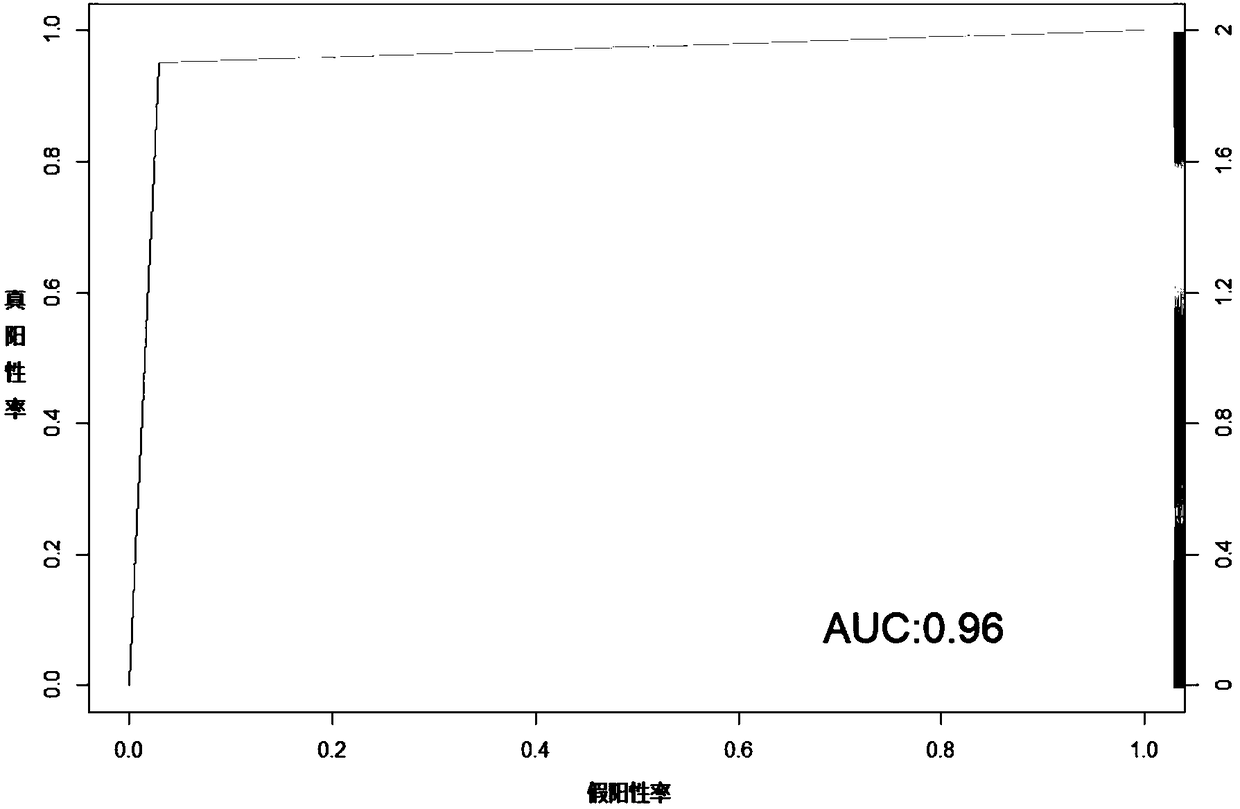
Dysfunctional or exhausted T cells arise in chronic diseases including chronic viral infections and cancer, and express high levels of co-inhibitory receptors. Therapeutic blockade of these receptors has clinical efficacy in the treatment of cancer. While co-inhibitory receptors are co-expressed, the triggers that induce them and the transcriptional regulators that drive their co-expression have not been identified. The immunoregulatory cytokine IL-27 induces a gene module in T cells that includes several known co-inhibitory receptors (Tim-3, Lag-3, and TIGIT). The present invention provides a novel immunoregulatory network as well as novel cell surface molecules that have an inhibitory function in the tumor microenvironment. The present invention further provides the novel discovery that the transcription factors Prdm1 and c-Maf cooperatively regulate the expression of the co-inhibitory receptor module. This critical molecular circuit underlies the co-expression of co-inhibitory receptors in dysfunctional T cells and identifies novel regulators of T cell dysfunction.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC +2
Multiple myeloma molecular subtype and application thereof to medication guidance
ActiveCN108559778AImprove understandingMicrobiological testing/measurementTherapeutic effectCancer research
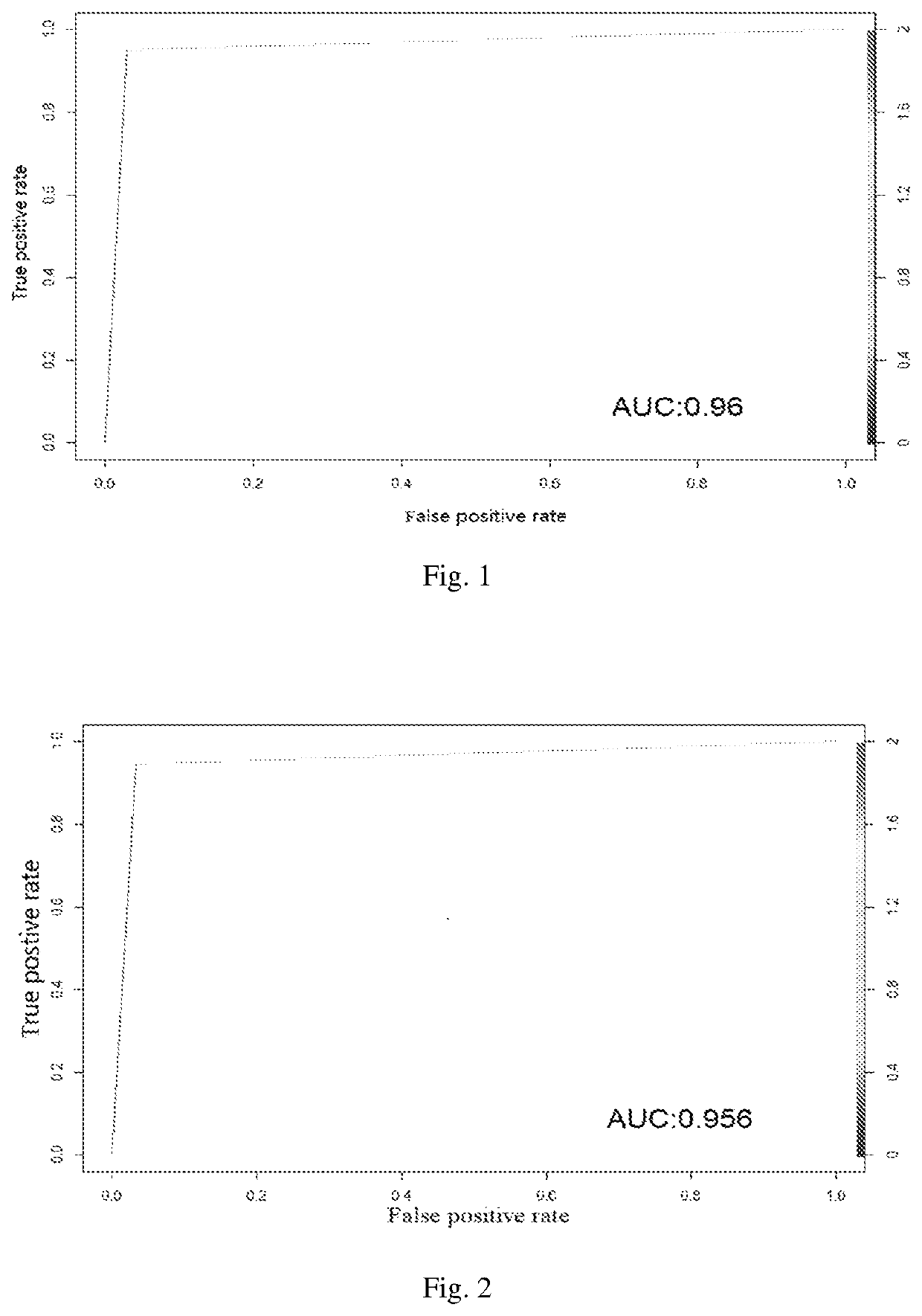
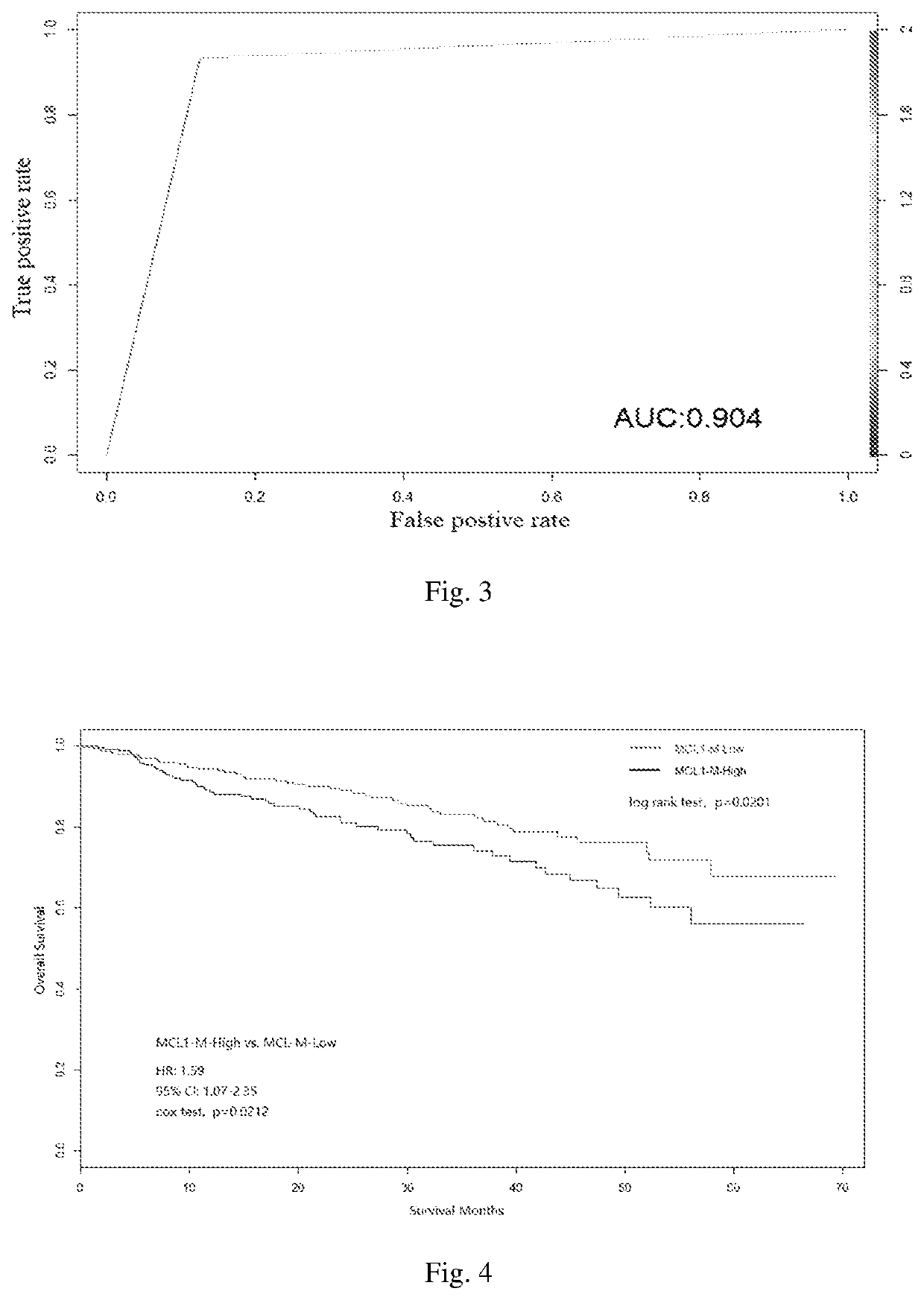
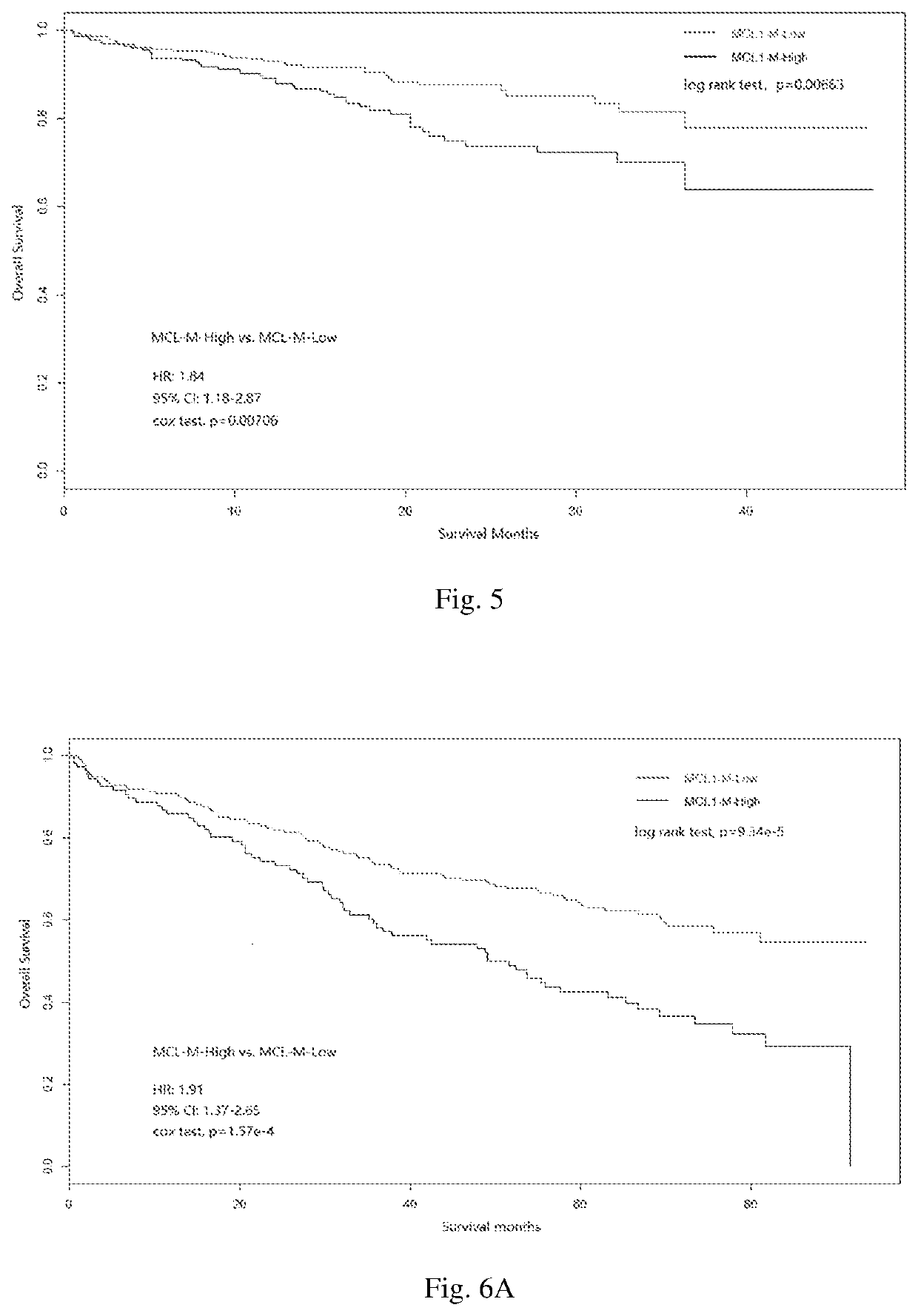
The invention discloses a multiple myeloma molecular subtype and the application thereof to medication guidance. The invention provides the application of a substance for acquiring or detecting expression of 97 genes in to-be-detected patients suffering from multiple myeloma tumor to preparation of a production for detecting the treatment effect of bortezomib or a bortezomib-containing medicine for detecting the to-be-detected patients suffering from multiple myeloma tumor. The invention identifies a gene module capable of co-expressing with an MCL1 gene (short for MCL1-M). The gene module isused for dividing multiple myeloma into two main subtypes such as an MCL-M-High subtype and an MCL-M-Low subtype. The two subtypes have obviously different prognostic and genetic features.
Owner:北京瑞牧西康医疗器械有限公司
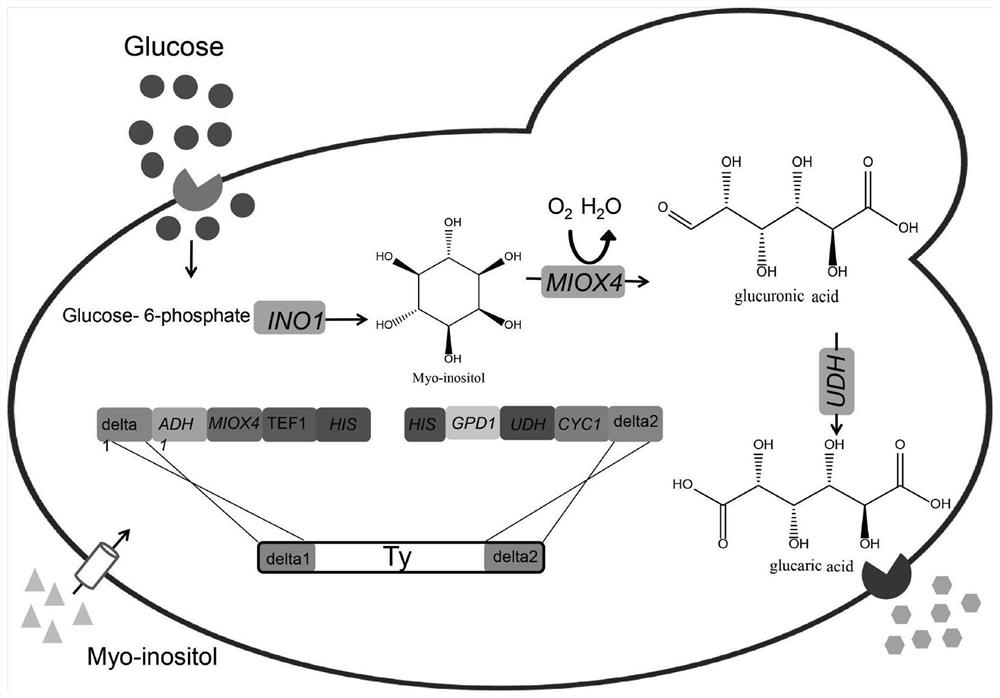
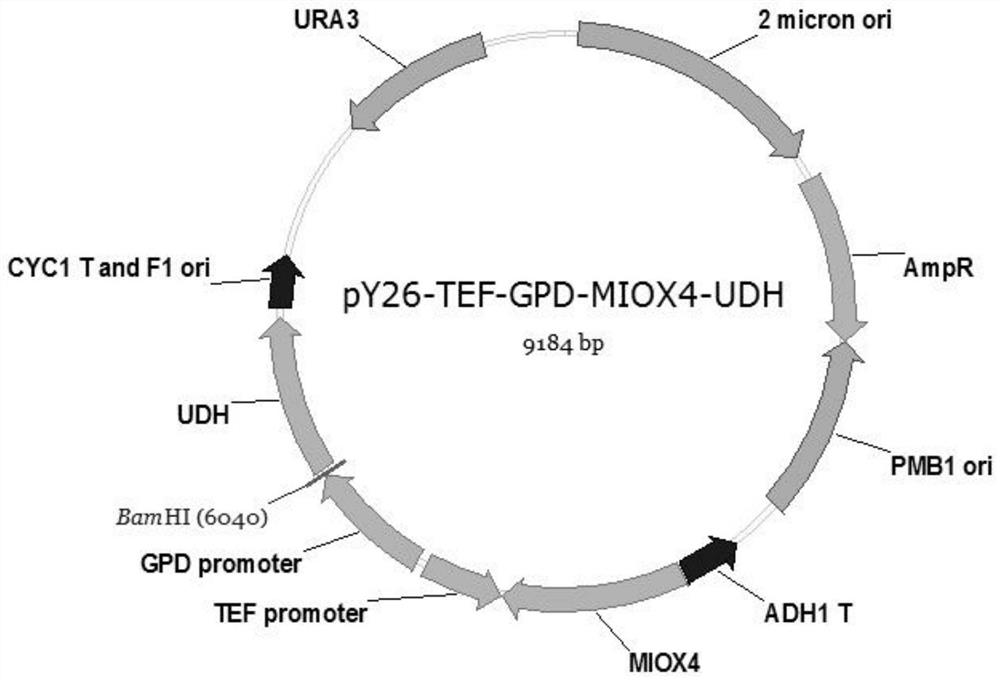
Method for producing glucaric acid by improving saccharomyces cerevisiae engineered strain fermentation
The invention discloses a method for producing glucaric acid by improving saccharomyces cerevisiae engineered strain fermentation, and belongs to the technical field of bioengineering. According to the method disclosed by the invention, Delta sites of multiple copies in a saccharomyces cerevisiae genome are used as exogenous gene module integration sites, so that one-time integration of an exogenous gene module is realized so as to fulfil the aim of expression of the multiple copies. According to the method disclosed by the invention, through an integration expression way, gene engineering bacteria subjected to the integration of genome Delta sites is passaged for 20 times; and under the same fermentation condition, the yield of the glucaric acid is still 3.8g / L, and the method has an important application meaning on industrial production of the glucaric acid.
Owner:JIANGNAN UNIV
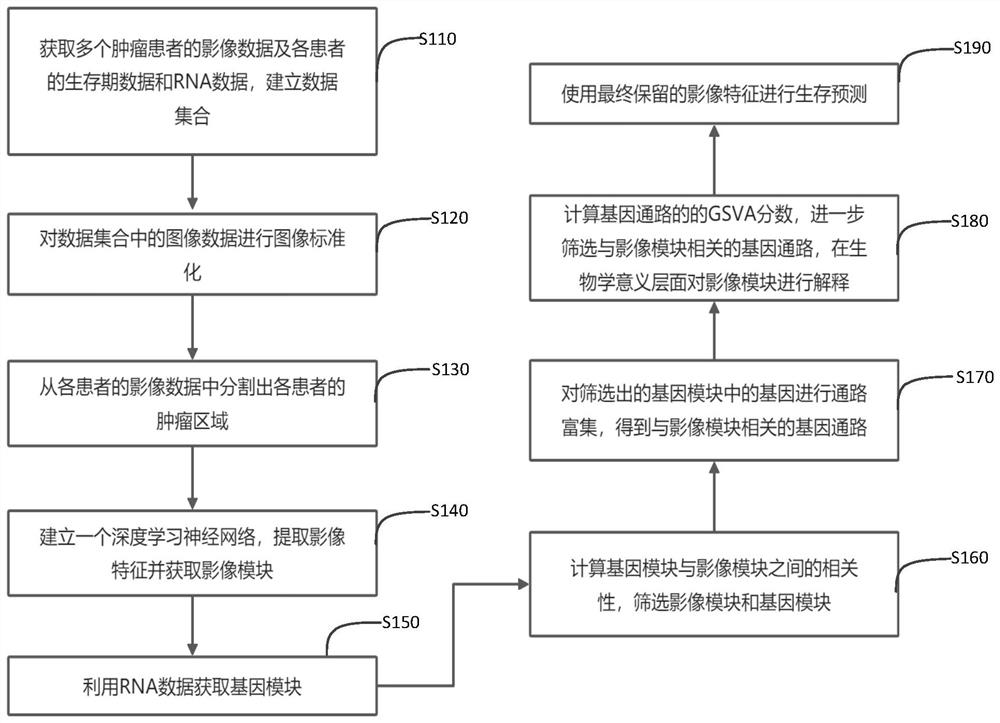
Survival prediction method and system based on image genomics
ActiveCN112907555AImprove generalization abilityImprove interpretabilityImage enhancementImage analysisData setComputational gene
The invention discloses a survival prediction method and system based on image genomics. The method comprises the following steps: acquiring image data of tumor patients and lifetime data and RNA data of each patient, and establishing a data set; segmenting a tumor area of each patient from the image data; inputting the image data of each patient into a neural network to extract image features and cluster the image features to obtain a plurality of image modules; obtaining a gene module of each patient by using the RNA data; performing screening according to the correlation between the gene modules and the image modules, and selecting a plurality of gene modules and image modules which are strongly correlated; performing pathway enrichment on genes in the selected gene module to obtain a gene pathway related to the image module; calculating a gene set variation analysis score of the gene pathway, and retaining the gene pathway having strong correlation with the image module; and carrying out survival prediction by using the retained image features. According to the invention, the biological interpretability in the aspect of survival prediction can be improved, and the generalization ability of deep learning can be improved at the same time.
Owner:SHENZHEN INST OF ADVANCED TECH CHINESE ACAD OF SCI
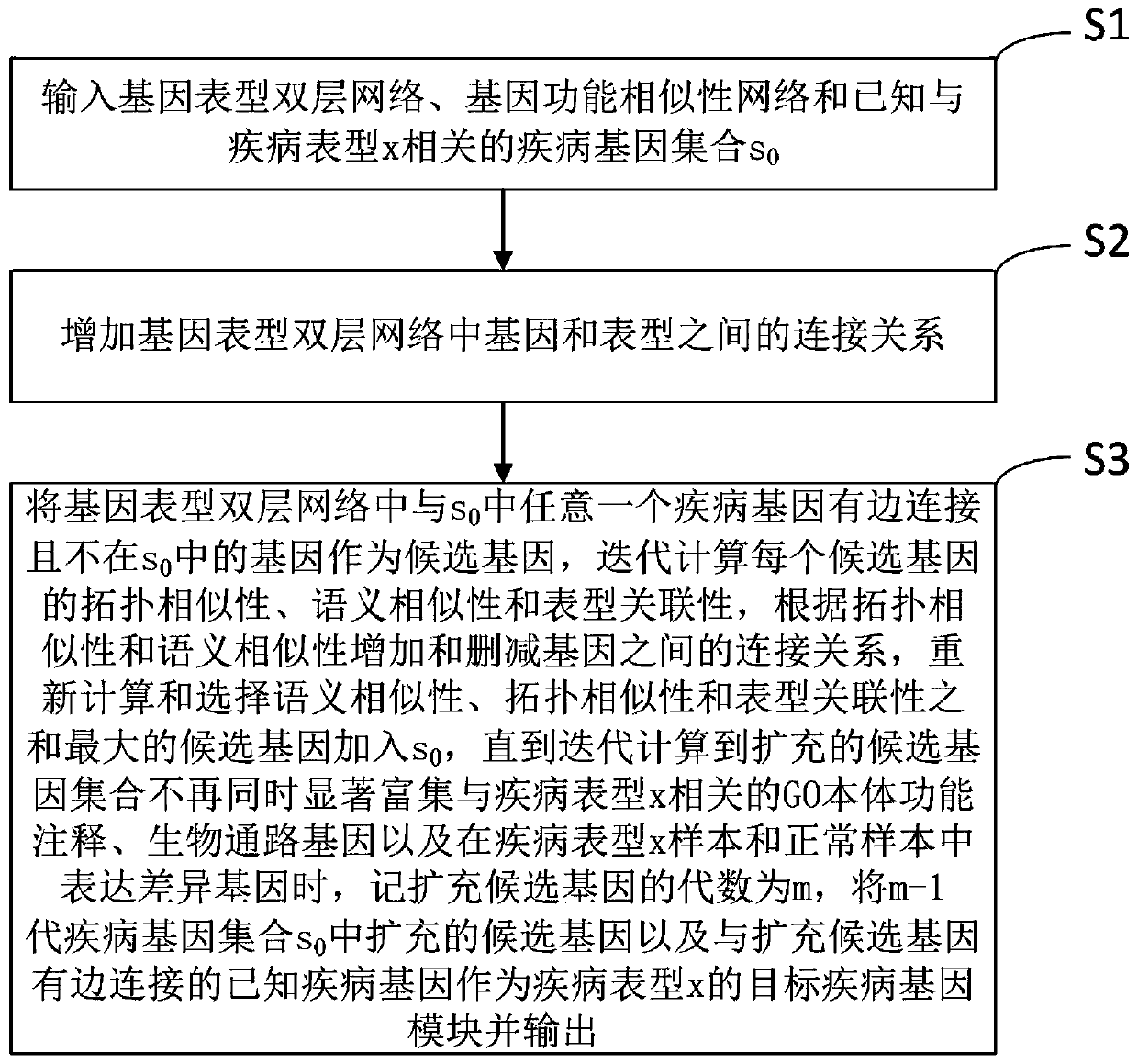
Gene module analysis method
The invention discloses a gene module analysis method. The gene module analysis method comprises the following steps: inputting a gene phenotype double-layer network, a gene function similarity network and a known disease gene set s0 related to a disease phenotype; increasing the connection relationship between genes and phenotypes in the gene phenotype double-layer network; taking genes which arein edge connection with disease genes in s0 and are not in s0 in the gene phenotypic double-layer network as candidate genes; calculating and selecting a candidate gene with the maximum sum of semantic similarity, topological similarity and phenotypic relevance, and adding the candidate gene into s0; and when the expanded candidate gene set does not significantly enrich the GO ontology function annotation and the biological pathway gene related to the disease phenotype at the same time and express the differential gene in the disease phenotype sample and the normal sample any more, recordingthe current algebra as m, and outputting m-1 as candidate genes expanded in the first generation s0 and known disease genes connected with the edges of the expanded candidate genes.
Owner:ANHUI UNIVERSITY
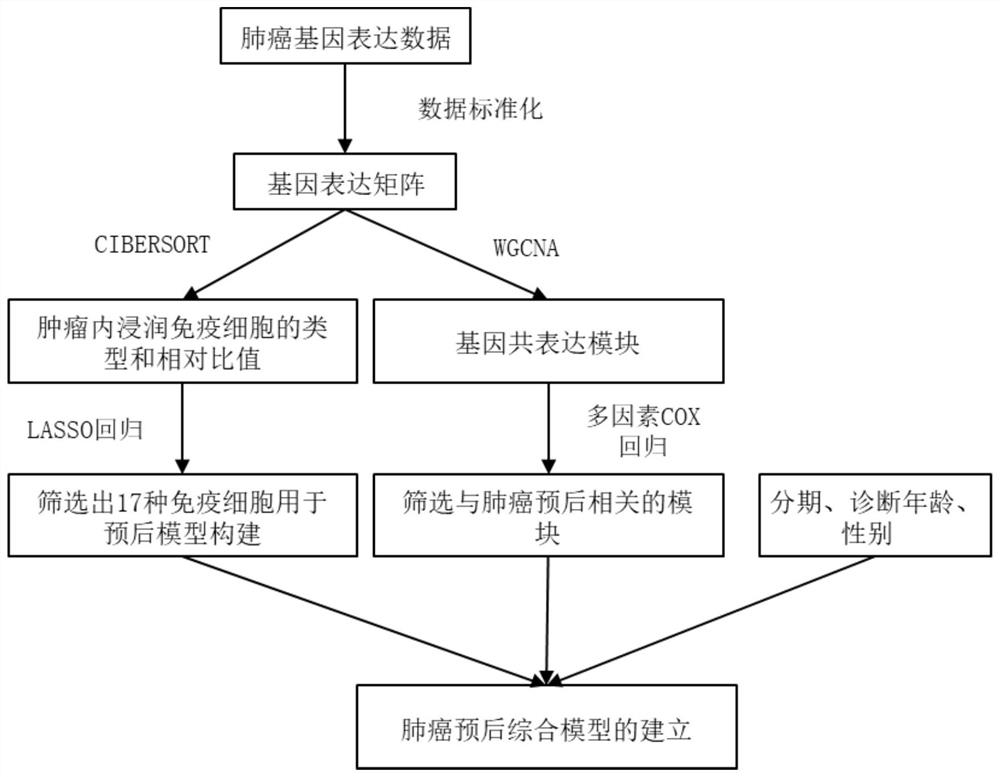
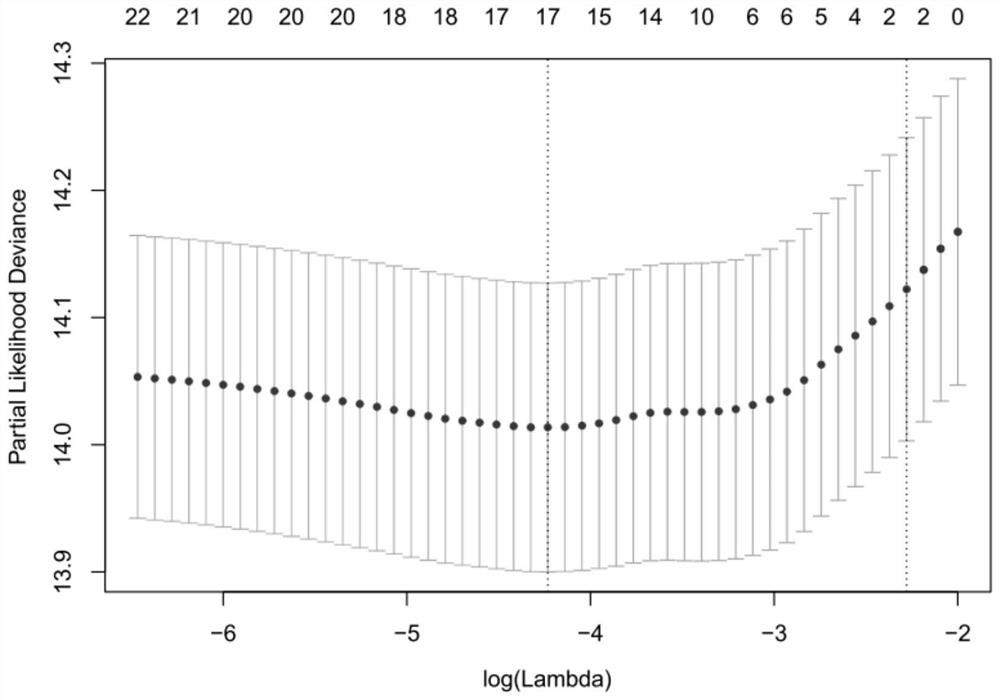
Lung cancer prognosis comprehensive prediction model, construction method and device
ActiveCN112635063AHigh clinical application valueMedical simulationMedical data miningOncologyBiology
The invention discloses a lung cancer prognosis comprehensive prediction model and a construction method and device. The method comprises the following steps: collecting original gene expression data and corresponding clinical survival data of a lung cancer sample and conducting data preprocessing and standardization, thereby acquiring a gene expression matrix; obtaining the types of immune cells in the tumor and calculating the relative ratio of the various types of immune cells; screening out parameters for constructing a prognosis prediction model from the obtained immune cell types and obtaining corresponding regression coefficients, wherein the parameters are multiple immune cell types; based on the screened parameters, calculating an immune score according to the relative ratio and the corresponding regression coefficient; identifying a gene co-expression module, searching for a gene module cooperatively expressed in the lung cancer sample, and determining a gene module related to prognosis; and constructing a prognosis comprehensive prediction model. According to the invention, immune scores, clinical information and gene co-expression module characteristics are integrated to construct a comprehensive prediction model to predict prognosis of lung cancer patients.
Owner:SOUTH CHINA UNIV OF TECH
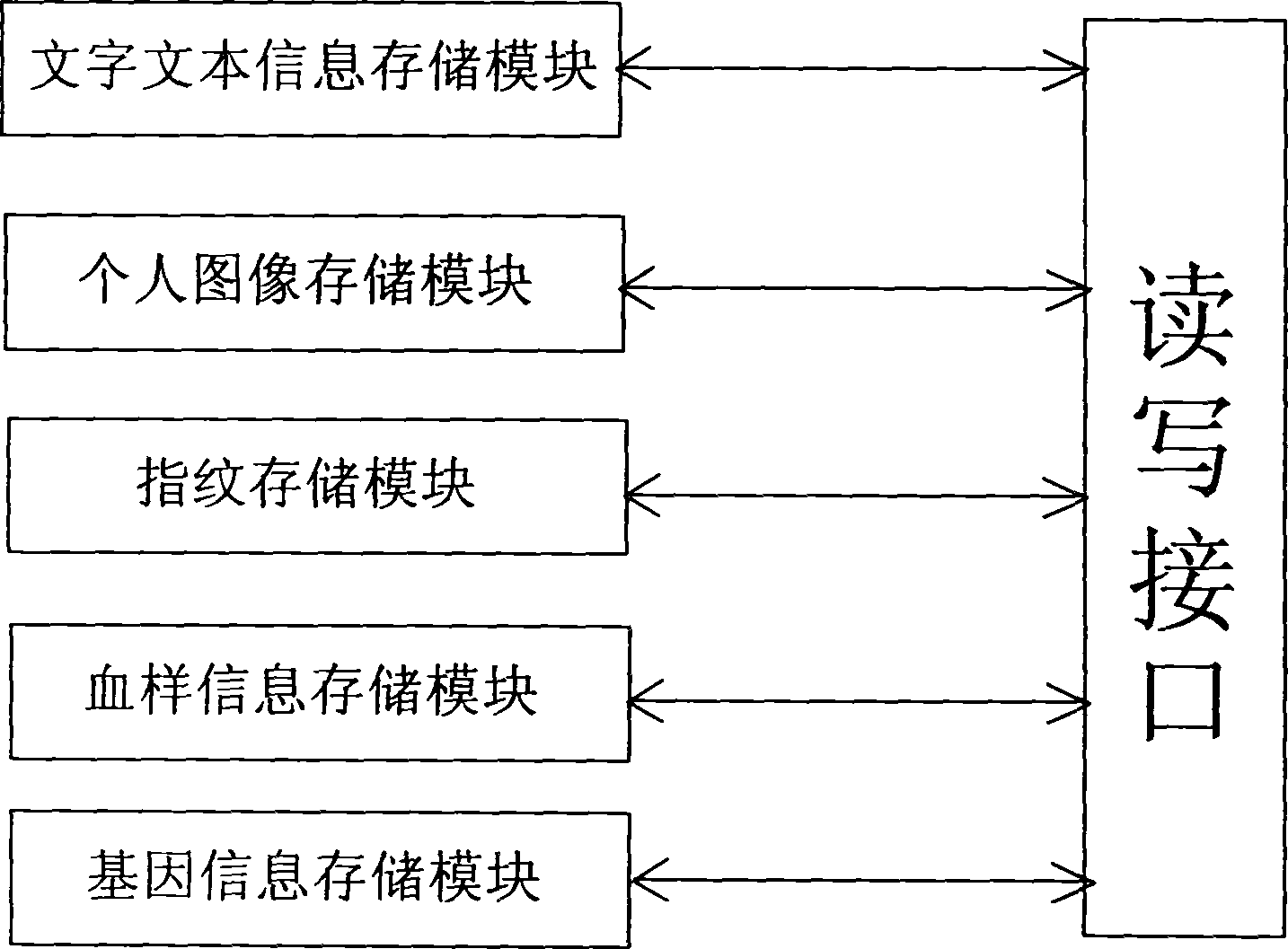
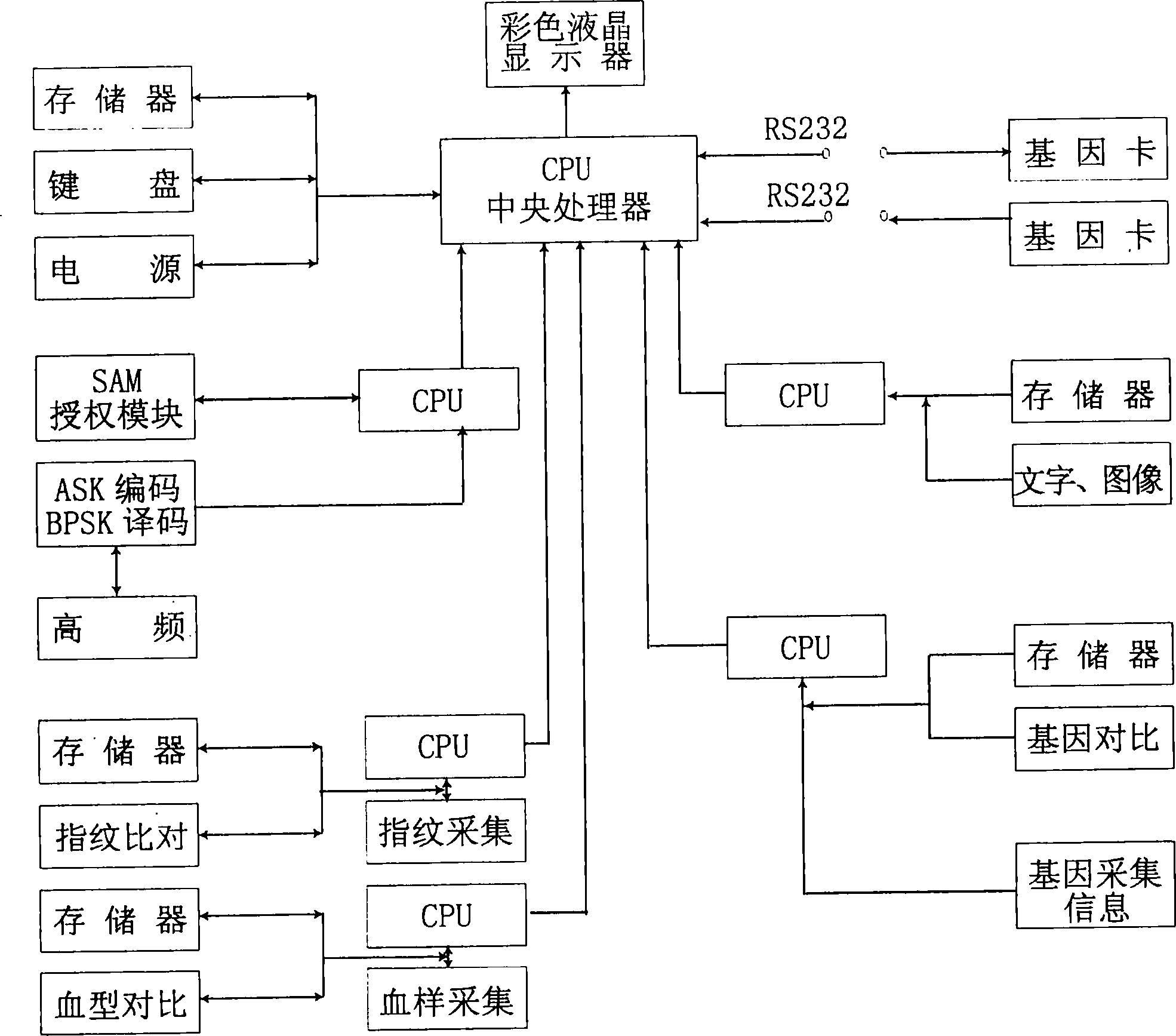
Gene health card and gene recorder
InactiveCN1889100AFacilitate active conversionImprove service qualitySensing record carriersRecord carriers used with machinesDna encodingApplication areas
The invention is about the gene health card and the gene recorder. The gene recorder includes the power supply, the operation control keyboard, the memorizer, the monitor, the CPU, the encryption / deciphering module, the fingerprint module, the blood type module, the character figure module, the gene module and the faucet which matches to the read-write interface of the gene health card. The fingerprint module and the encryption / deciphering module identify the personal identity then change it into the electronic number such as the DNA detecting results(the personal DNA encoding and the atlas) and memory it to transfer to other databank or the personal gene health card. The gene health card records the personal stature, the weight, the age, the sex, the head portrait, the blood type, the fingerprint, the DNA encoding and the part DNA atlas, so it is benefit for analyzing the personal health.
Owner:扬州金鼎电子有限公司
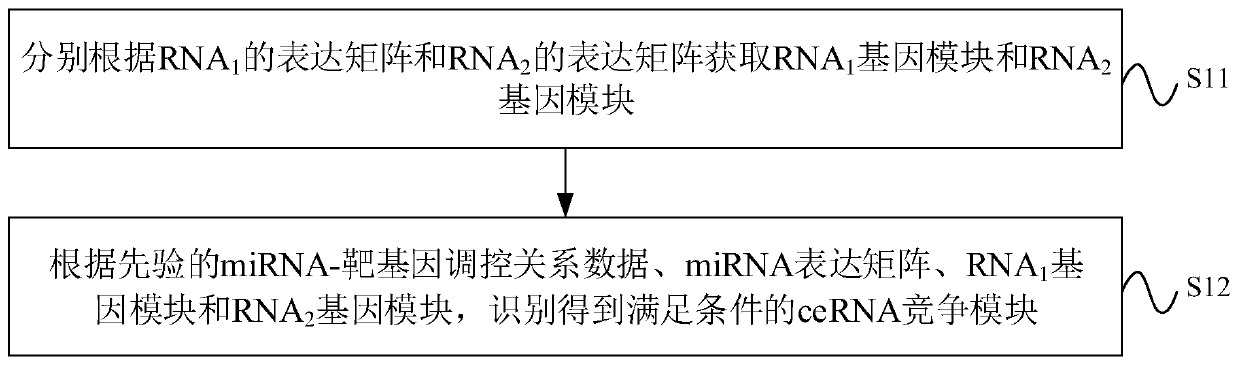
CeRNA competition model identification method and device, electronic equipment and storage medium
ActiveCN111383709AAccurate identificationCharacter and pattern recognitionProteomicsAlgorithmEngineering
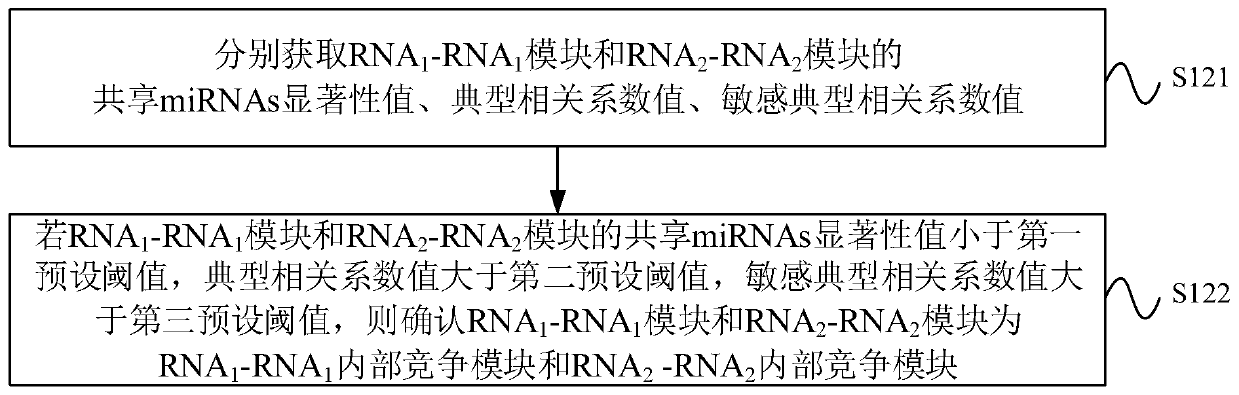
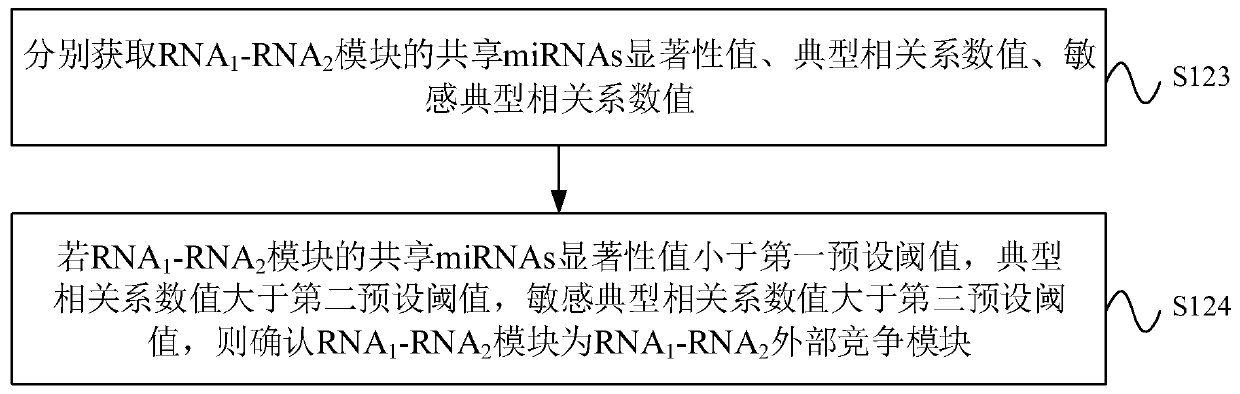
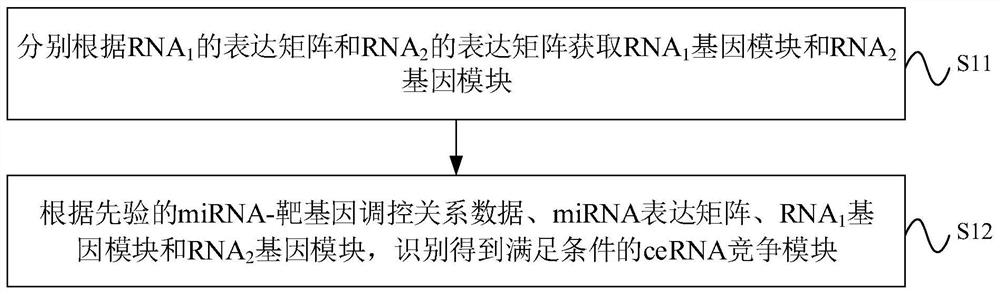
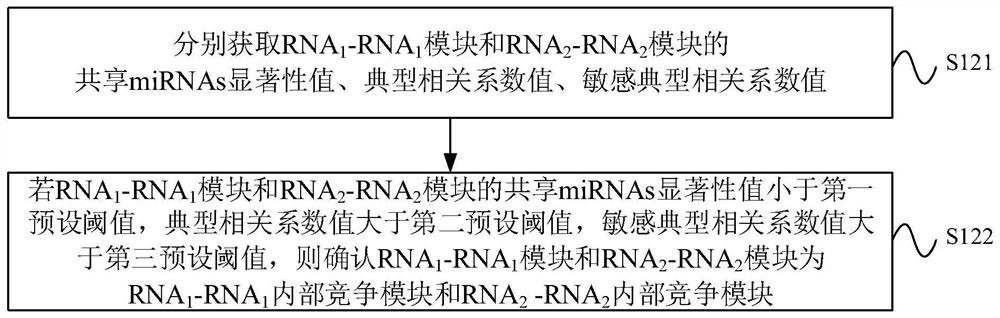
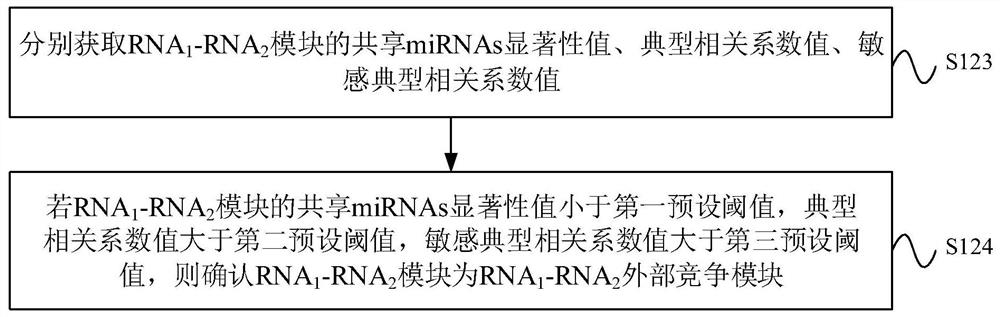
The invention provides a ceRNA competition model identification method and device, electronic equipment and a storage medium, and relates to the field of gene identification. The ceRNA competition model identification method comprises the steps of obtaining an RNA1 gene module and an RNA2 gene module according to the expression matrix of the RNA1 and the expression matrix of the RNA2, wherein theRNA1 and the RNA2 are miRNA target genes; regulating relation data, a miRNA expression matrix, an RNA1 gene module and an RNA2 gene module according to the prior miRNA-target gene, and performing identification for obtaining the ceRNA competition module which satisfies a condition, wherein the ceRNA competition module comprises an inner competition module and an outer competition module. Because the inner and outer competition conditions of the ceRNA are considered, the inner and outer competition strength of the ceRNA and the influence of the miRNA to the inner and outer competition strengthsof the ceRNA are measured in a module level. Therefore, the method provided by the invention can accurately identify the ceRNA competition module.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA +1
Multiple myeloma molecular subtype and application
ActiveCN108570501AImprove understandingMicrobiological testing/measurementBiostatisticsCancer researchGene expression
The invention discloses a multiple myeloma molecular subtype and application. The product provided by the invention comprises a substance for obtaining or detecting 97 gene expressions in a multiple myeloma cancer patient to be detected. The product further comprises equipment for operating a multiple myeloma Bayes classifier. In the invention, a gene module (MCL1-M for short) co-expressed with anMCL1 gene is identified, and the gene module is applied to divide multiple myeloma into two main subtypes, namely an MCL-M-High subtype and an MCL-M-Low subtype. The two subtypes have prognosis and genetics characteristics which are significantly different.
Escherichia coli recombinant bacterium for denovo synthesis of vitamin B12, and construction method and application of escherichia coli recombinant bacterium
The invention provides an escherichia coli recombinant bacterium for denovo synthesis of a vitamin B12, and a construction method and application of the escherichia coli recombinant bacterium. Specifically, escherichia coli serves as a starting strain, the engineering bacterium containing an HBA gene module, an HBAD gene module, a CBAD gene module and a Cbi gene module is constructed, the engineering bacterium efficiently and rapidly produces the B12 (the yield is at least 50 times higher than that of the prior art), and simple compounds can even be used as raw materials to conduct denovo synthesis on the vitamin B12.
Owner:TIANJIN INST OF IND BIOTECH CHINESE ACADEMY OF SCI
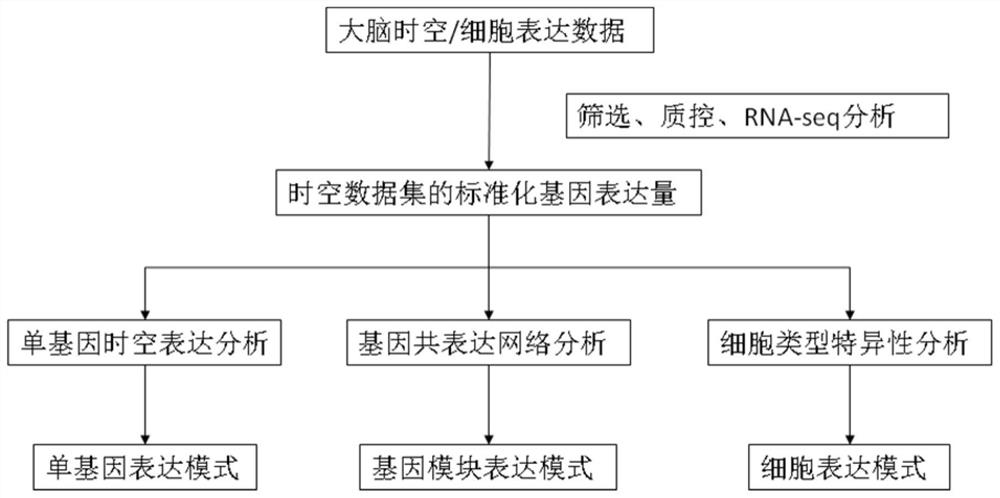
Method for establishing human brain gene expression space-time norm
ActiveCN110349625AConvenient supplementEasy to compareBiostatisticsHybridisationData setOriginal data
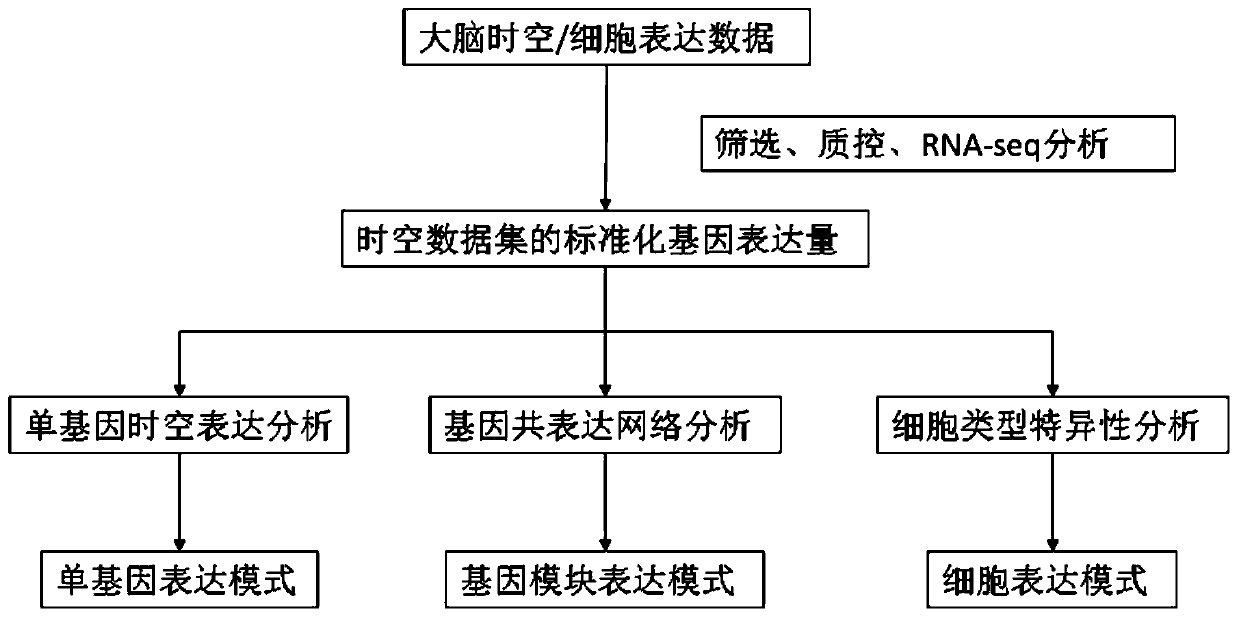
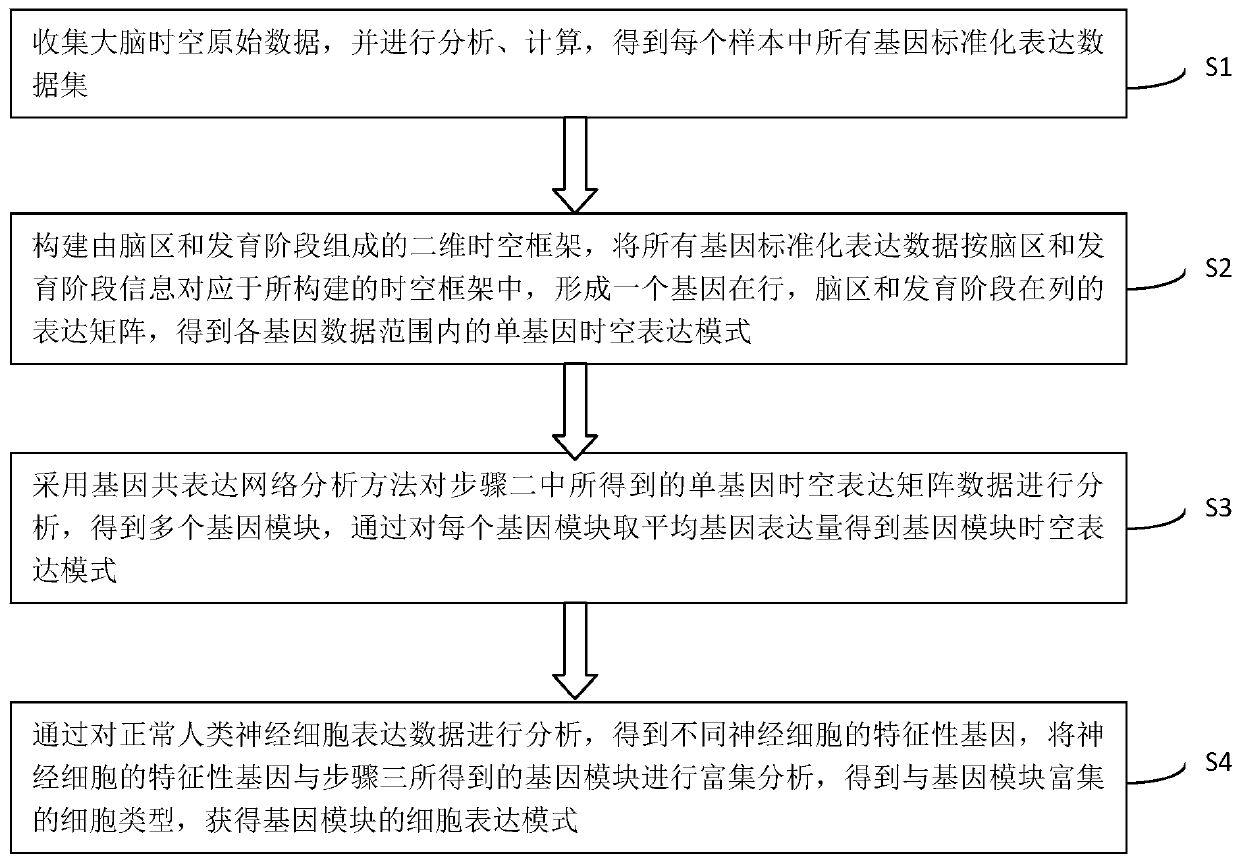
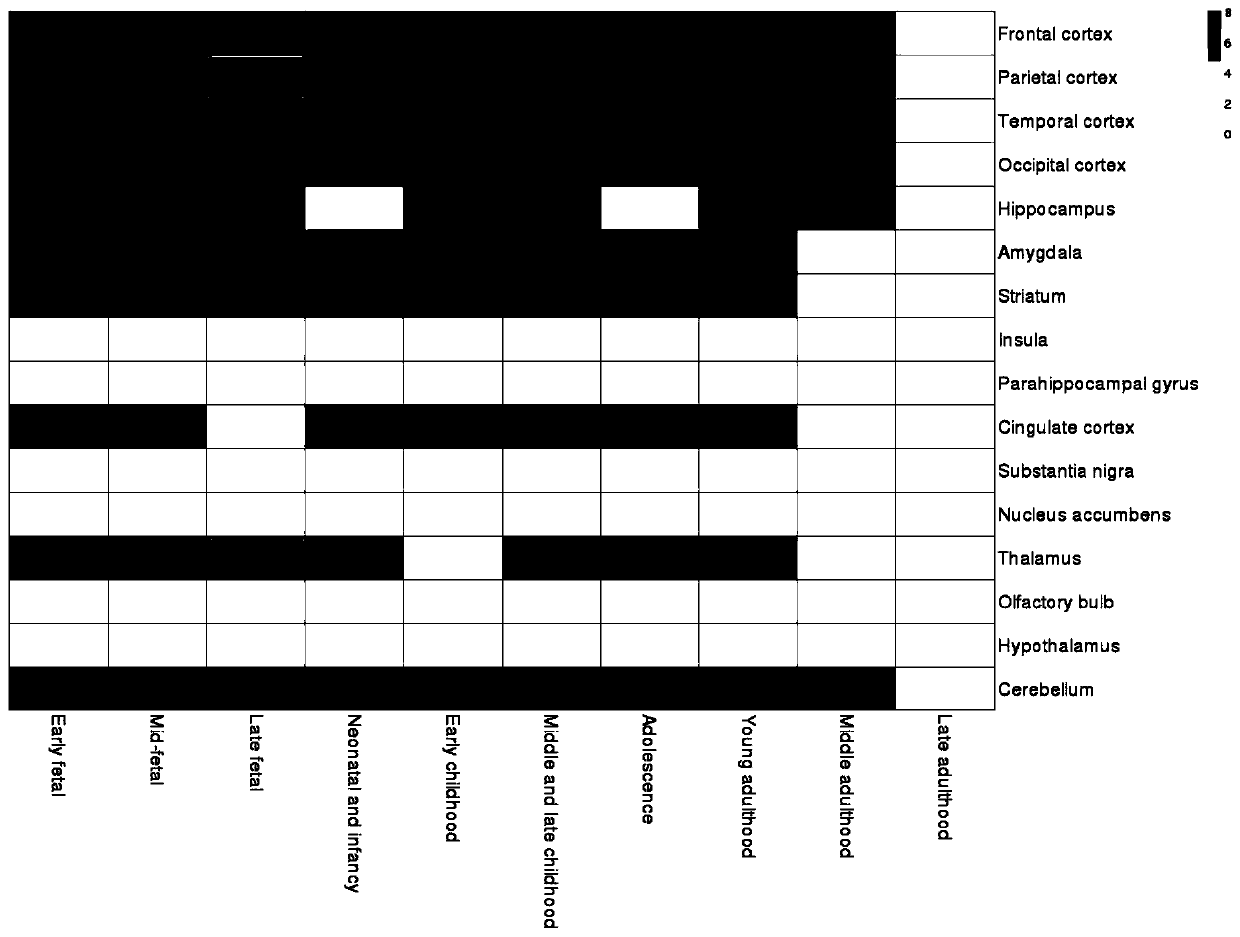
The invention discloses a method for establishing a human brain gene expression space-time norm. The method comprises the following steps: collecting brain space-time original data to obtain a gene standardized expression data set; constructing a two-dimensional space-time framework of a brain region and a development stage, and enabling all gene standardized expression data to correspond to the constructed space-time framework according to the brain region and development stage information to obtain a single-gene space-time expression mode; analyzing the obtained single-gene space-time expression matrix data by adopting a gene co-expression network analysis method to obtain a plurality of gene modules, and averaging the gene expression quantities of the gene modules to obtain a gene module space-time expression mode; analyzing the normal human nerve cell expression data, and performing the enrichment analysis of characteristic genes of nerve cells and the obtained gene modules to obtain a cell expression mode of the gene module. According to the method, all reference data sets are finally presented in the same two-dimensional space-time framework, so that the data sets from different sources are integrated, and researchers can intuitively supplement and compare the content of the space-time framework.
Owner:INST OF PSYCHOLOGY CHINESE ACADEMY OF SCI
Adenoviral assembly method
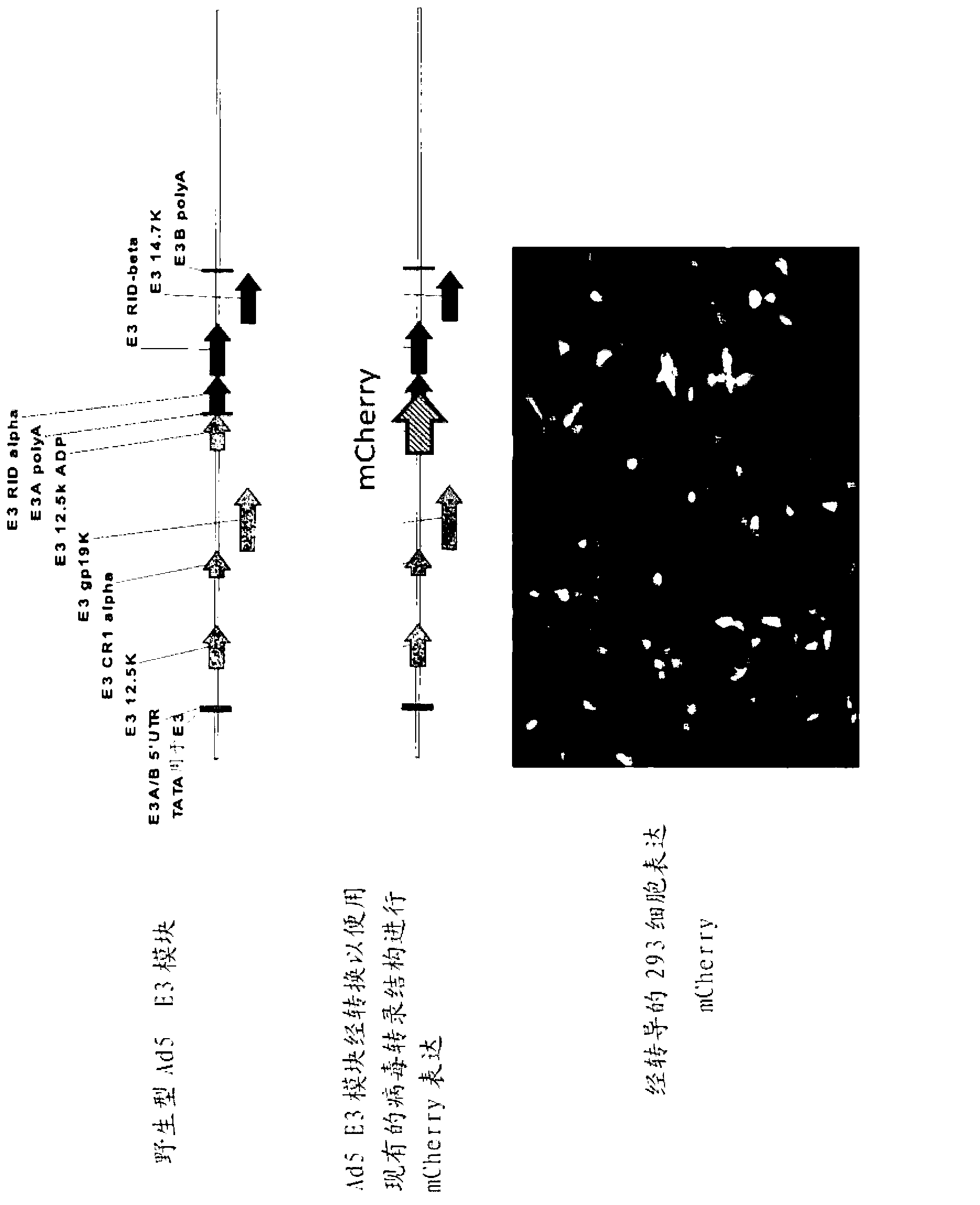
Methods of assembling modified adenoviruses, libraries of adenoviral gene modules and compositions thereof are provided herein.
Owner:SALK INST FOR BIOLOGICAL STUDIES
Molecular typing of multiple myeloma and application
PendingUS20210055301A1Predict riskHealth-index calculationMicrobiological testing/measurementMedicineMCL1
Disclosed are molecular typing of multiple myeloma and application thereof. Specifically, disclosed is a product comprising a substance for obtaining or detecting 97 gene expressions in multiple myeloma patients to be detected and an apparatus for operating a multiple myeloma Bayesian classifier. By using the product, the present invention identifies a gene module co-expressed with the MCL1 gene, thereby distinguishing molecular subtypes of multiple myeloma having different prognoses and bortezomib sensitivities.
Owner:BEIJING NORMAL UNIVERSITY
Method for screening transcription factors related to postharvest softening of grape fruits
InactiveCN114107456AMicrobiological testing/measurementProteomicsBiotechnologyH2O2 - Hydrogen peroxide
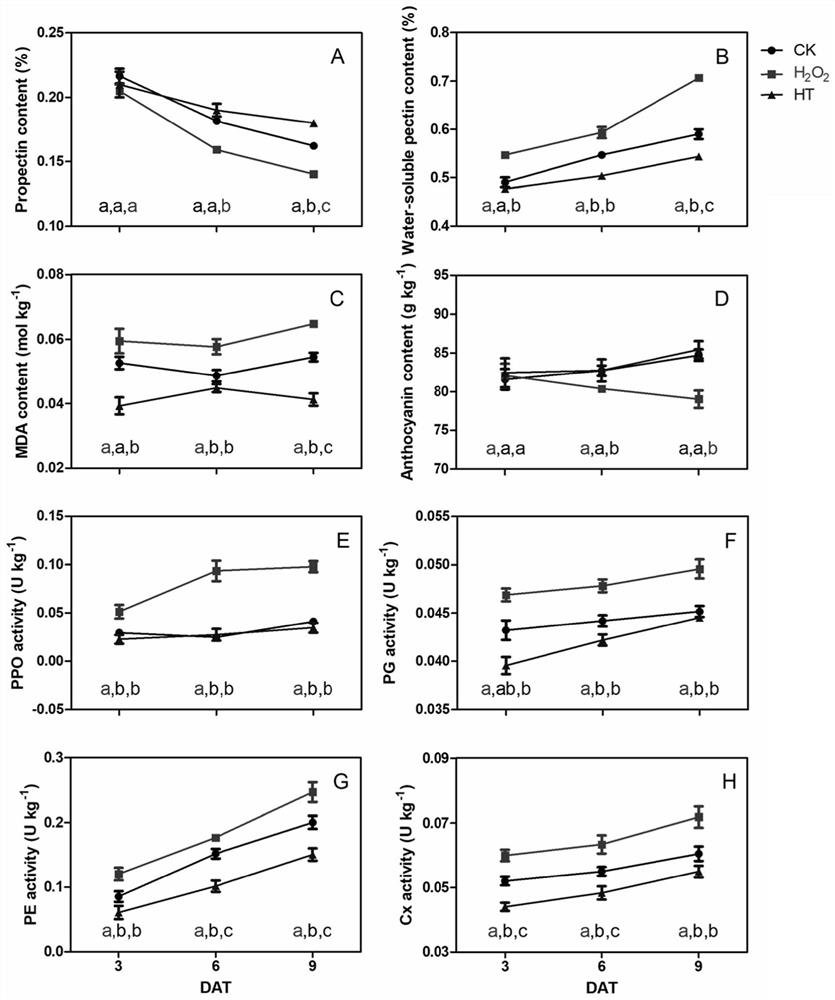
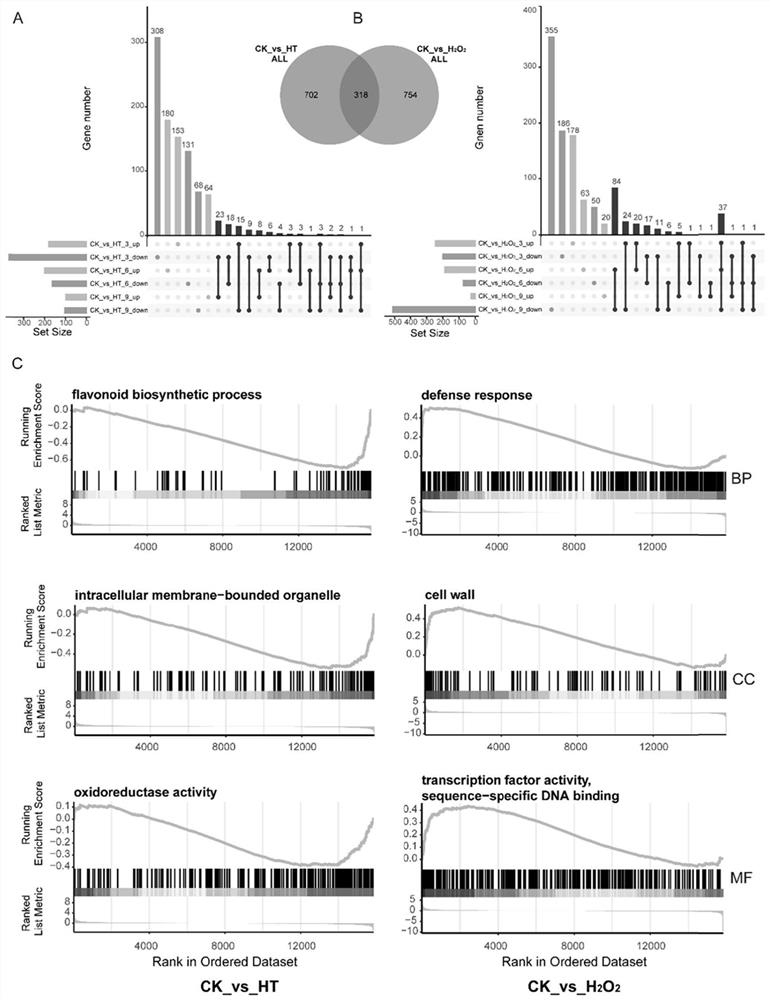
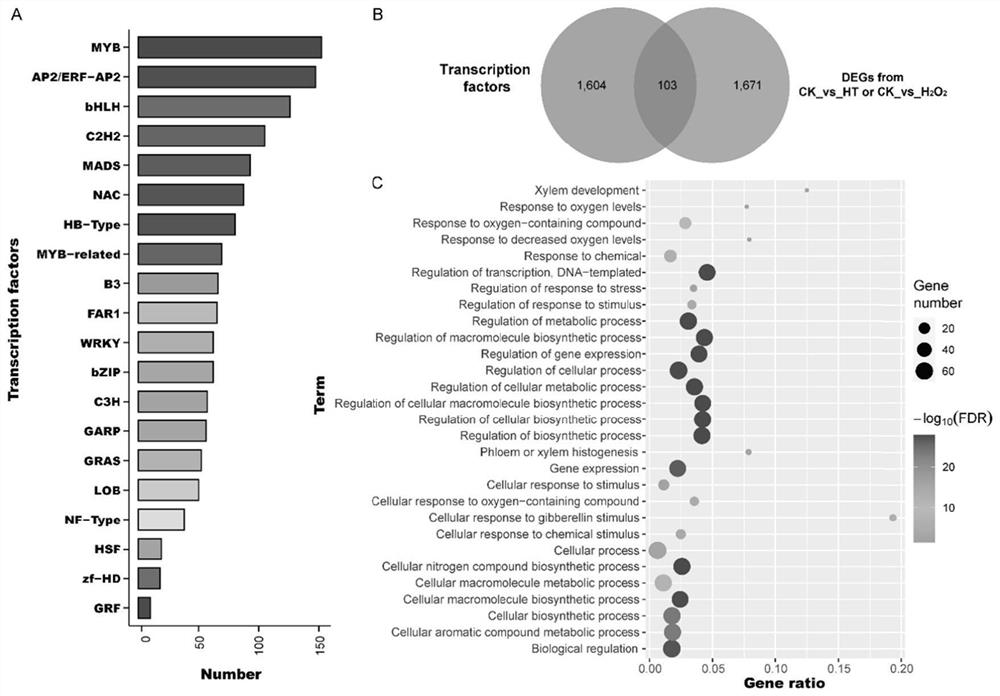
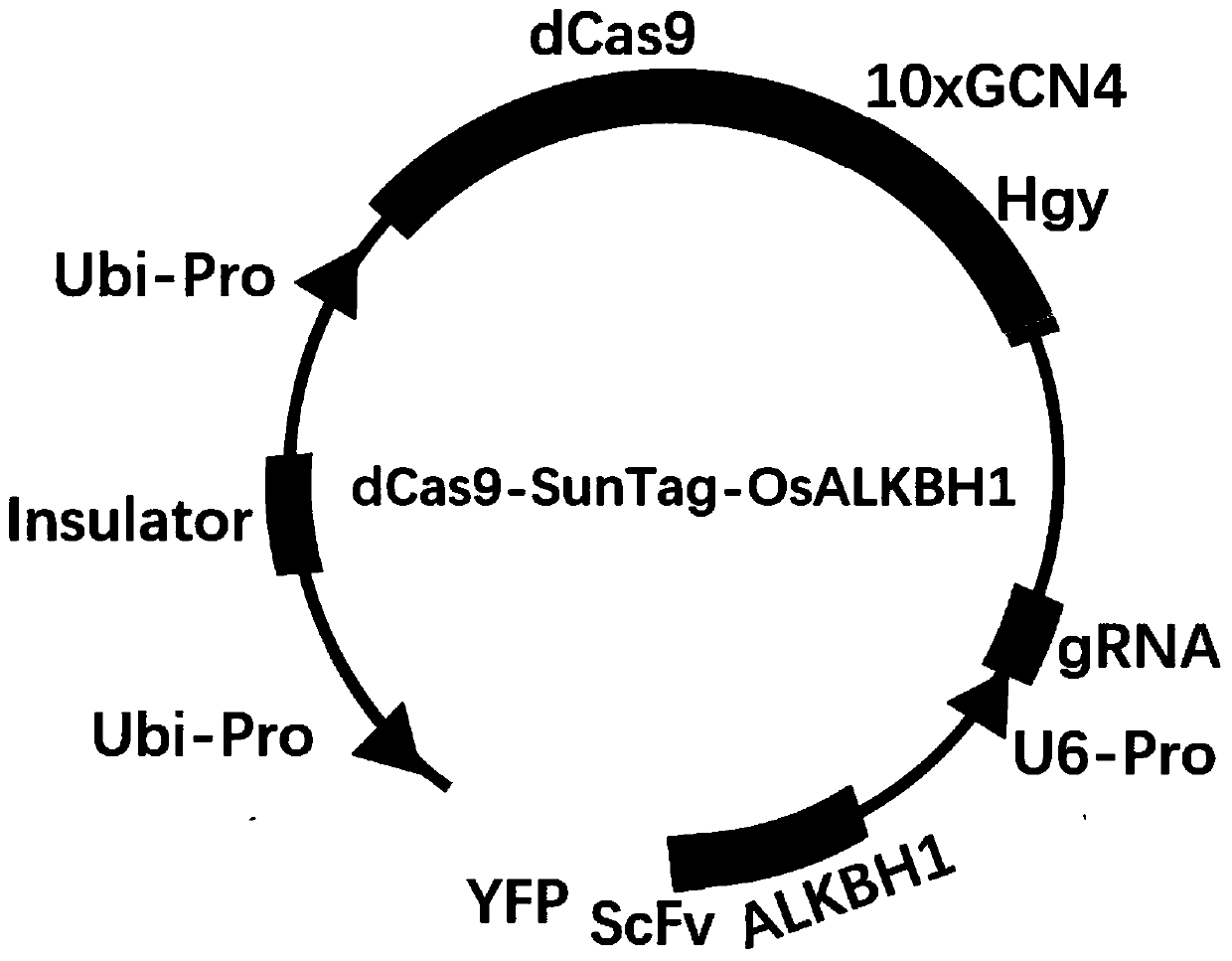
The invention belongs to the technical field of high-throughput sequencing data analysis, and particularly relates to a method for screening transcription factors related to postharvest softening of grape fruits. According to the invention, hypotaurine (HT) and hydrogen peroxide (H2O2) treatment is carried out on harvested 'Kyoho' grapes, through transcriptome sequencing and weighted gene co-expression network analysis (WGCNA), correlation analysis is carried out on differential expression genes obtained through transcriptome analysis and pectin content change, a differential expression gene module closely related to the pectin content change is screened out, and the specific value of the differential expression genes and the specific value of the pectin content change are obtained. And excavating differential expression transcription factors in the softening process of the picked grape fruits. According to the method, the softening character related transcription factors can be effectively screened, and a theoretical reference is provided for researching the softening occurrence mechanism of the picked grape fruits.
Owner:HENAN UNIV OF SCI & TECH
Method for efficiently converting gene module and line for rice leaf sheath protoplast
InactiveCN110656126AIncrease concentrationOptimizing Improvement Conversion TimeVector-based foreign material introductionPlant cellsBiotechnologyCentrifugation
The invention discloses a method for efficiently converting a gene module and line for a rice leaf sheath protoplast, and relates to a conversion method of the rice leaf sheath protoplast. The methodcomprises the steps of adding an MMg solution to the rice leaf sheath protoplast to obtain a rice leaf sheath protoplast suspension; adding plasmid and a polyethylene glycol-calcium solution of whichthe percentage by mass is 40% to the rice leaf sheath protoplast suspension, performing uniform mixing, and performing standing; and adding a W5 solution, performing uniform mixing, performing centrifugation, removing supernatant, performing suspension once again and precipitation on the W5 solution, performing centrifugation, removing supernatant, adding carbenicillin, performing uniform mixing,and performing culturing to complete transformation of the protoplast. According to the conversion method of the rice leaf sheath protoplast, preparation, plasmid concentration, conversion time and the like of the PEG solution are adjusted, so that the conversion efficiency is improved to 80%, intelligent reformation devices can exert functions in 80% of plant cells, available data can be providedfor research on functions of the devices at the initial stage, and convenience is provided for research on functions of the intelligent reformation devices for crops at the later stage.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
Efficient artificial rhizosphere combined nitrogen fixation system
ActiveCN111925974AImprove efficiencyGood growth promoting effectBacteriaGenetically modified cellsBiotechnologySynthetic biology
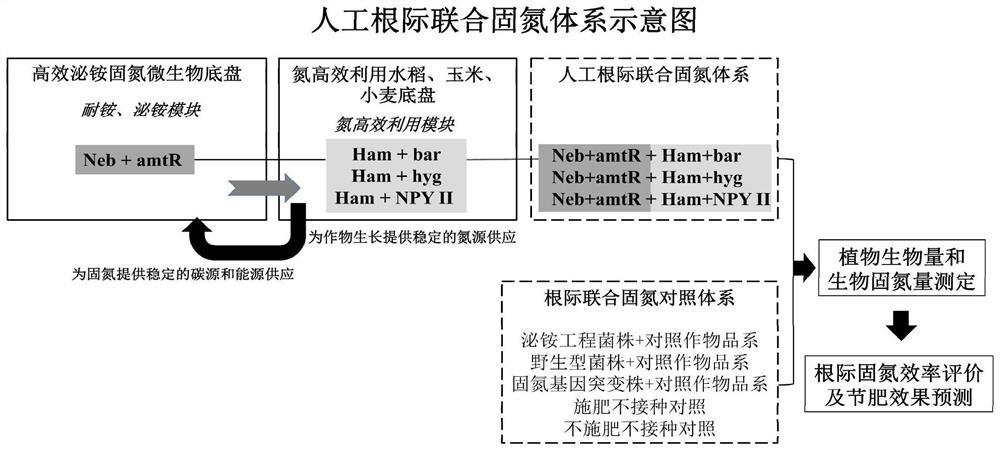
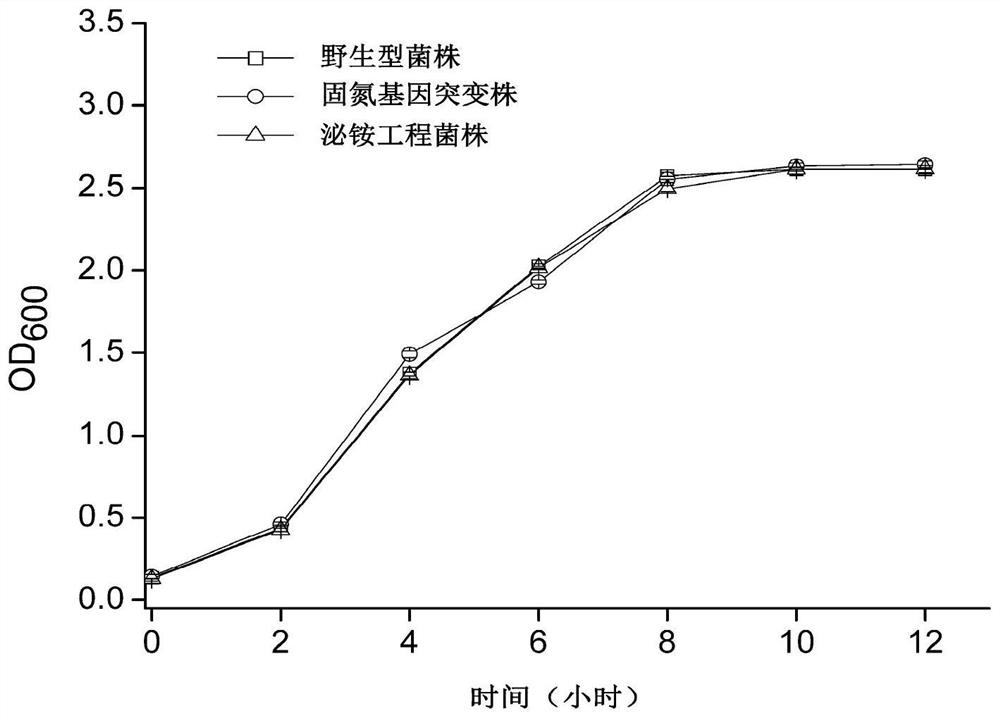
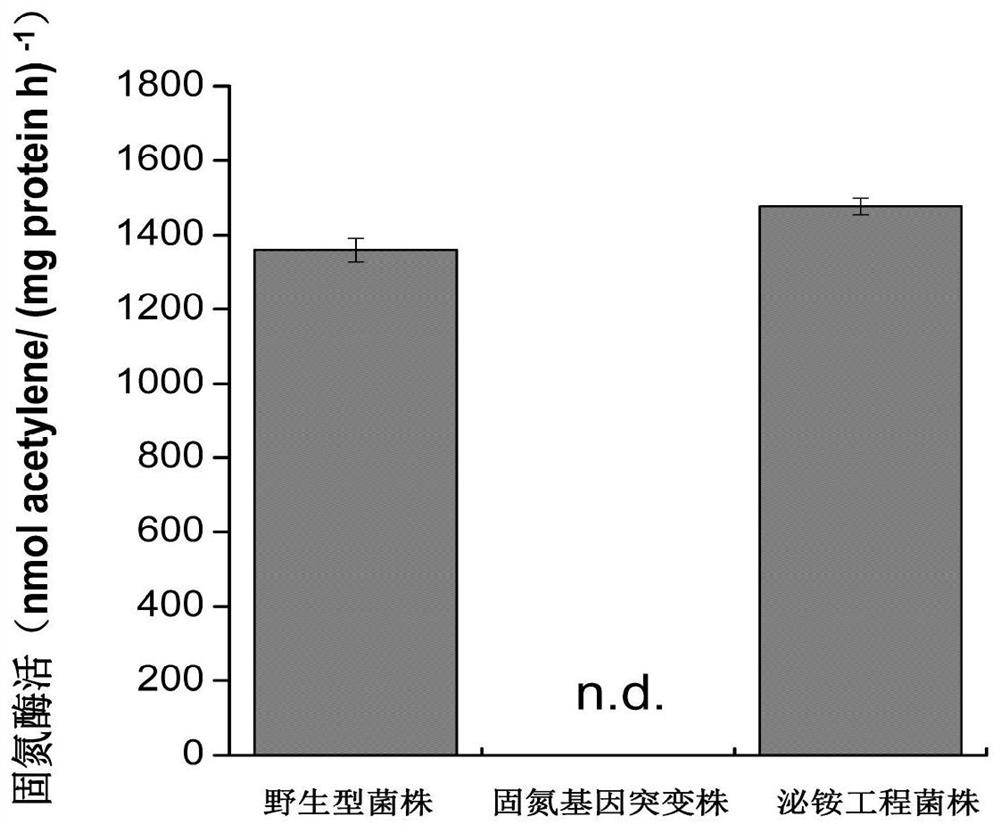
The invention provides an efficient artificial rhizosphere combined nitrogen fixation system constructed according to synthetic biology theories and methods. Two brand-new functional modules are respectively and manually designed: a'Neb + amtR 'ammonium secreting gene module is constructed in a nitrogen-fixing microorganism chassis, a'Ham' nitrogen efficient utilization module is constructed in non-leguminous crop chassis of rice, corn, wheat and the like, and functional coupling of the two artificial modules is realized through inoculation technologies such as seed coating and the like at crop rhizosphere. Compared with a combined nitrogen fixation system in the prior art, the artificial rhizosphere combined nitrogen fixation system has higher biological nitrogen fixation efficiency and plant growth promoting effect.
Owner:北京绿氮生物科技有限公司
Construction of mercury ion microorganism whole-cell biosensor taking violacein as output signal and application thereof
PendingCN113234651AAchieve qualitativeEasy to detectBacteriaMaterial analysis by observing effect on chemical indicatorOrganosolvA-DNA
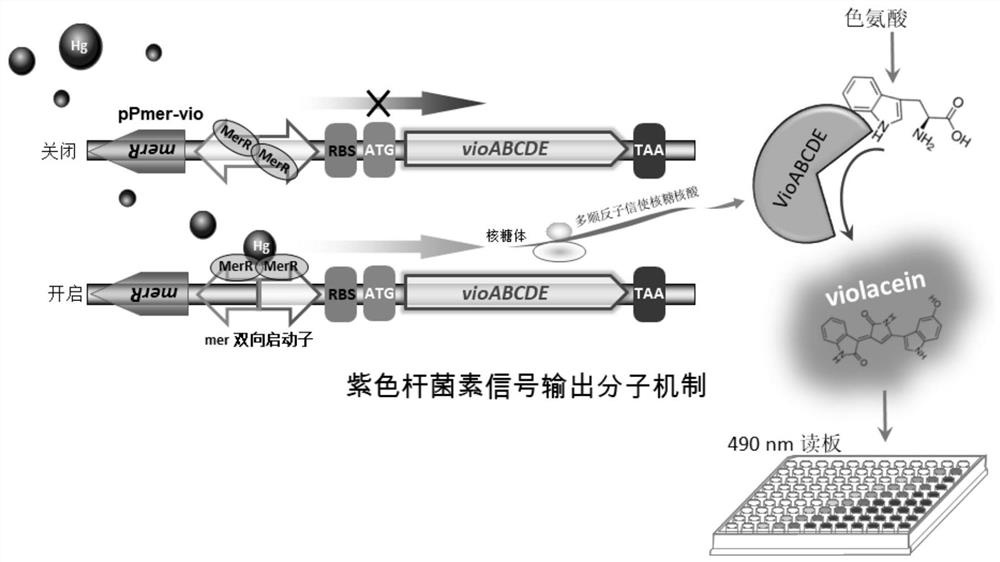
The invention discloses construction of a mercury ion microorganism whole-cell biosensor taking violacein as an output signal and an application thereof. The invention provides a recombinant bacterium which is obtained by introducing a recombinant vector A into a recipient bacterium, the recombinant vector A contains a DNA fragment A; the DNA fragment A contains a mer bidirectional promoter, one side of the mer bidirectional promoter is an MerR protein coding gene, and the other side of the mer bidirectional promoter is a violacein synthesis gene module. The recombinant bacterium provided by the invention can be used as a mercury ion microbial whole-cell biosensor taking violacein as an output signal. On one hand, qualitative indication of exposure of the target heavy metal can be achieved through change of cell colors, and on the other hand, intracellular violacein can be extracted through an organic solvent, and quantitative analysis of the target heavy metal is conducted through colorimetry.
Owner:SHENZHEN PREVENTION & TREATMENT CENT FOR OCCUPATIONAL DISEASES
Gene expression data analysis method based on PAM clustering algorithm
PendingCN113380326AEasy to digEasy to analyzeBiostatisticsCharacter and pattern recognitionAlgorithmGene recognition
The invention discloses a gene expression data analysis method based on a PAM clustering algorithm, and relates to the field of data analysis. The method comprises the steps of data acquisition, data preprocessing, gene module identification, GO enrichment analysis, PPI network construction, HUB gene identification and HUB gene verification. According to the method, on the basis of fully utilizing information contained in gene expression data, the optimal membership module can be searched for each gene through multiple iterations, so that the identified gene module is more reliable. The hidden information contained in the gene module can be better mined, so that the bioinformatics problem to be solved is comprehensively analyzed. According to the method, data preprocessing is performed on the gene expression data, so that the problems of much noise, many irrelevant genes, sparse data and the like in the gene expression data are solved. Through the downstream bioinformatics analysis process, a series of bioinformatics analysis can be completed, and the bioinformatics problems to be solved can be comprehensively analyzed and explained.
Owner:吉林省蒲川生物医药有限公司
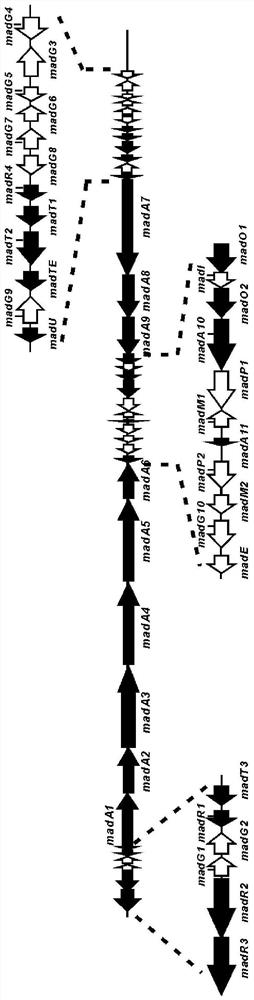
Biosynthetic gene cluster of maduramicin compound and application of biosynthetic gene cluster
The invention discloses a biosynthetic gene cluster of a maduramicin compound and application of the biosynthetic gene cluster. The biosynthetic gene cluster of the maduramicin compound comprises an I-type linear polyketone synthetase gene module, a glycosyl synthesis related gene module, a modified gene module and a regulatory gene module, and the I-type linear polyketone synthetase gene module comprises a madA1 gene, a madA2 gene, a madA3 gene, a madA4 gene, a madA5 gene, a madA6 gene, a madA7 gene, a madA8 gene, a madA9 gene, a madA10 gene and a madA11 gene; the glycosyl synthesis related gene module comprises a madG1 gene, a madG2 gene, a madG3 gene, a madG4 gene, a madG5 gene, a madG6 gene, a madG7 gene and a madG8 gene; the modified gene module comprises a madO1 gene, a madO2 gene, amadI gene, a madE gene, a madP1 gene, a madP2 gene, a madM1 gene, a madM2 gene, a madG9 gene, a madG10 gene and a madTE gene; and the regulatory gene module comprises a madR1 gene, a madR2 gene, a madR3 gene and a madR4 gene.
Owner:WUHAN HESHENG TECH CO LTD
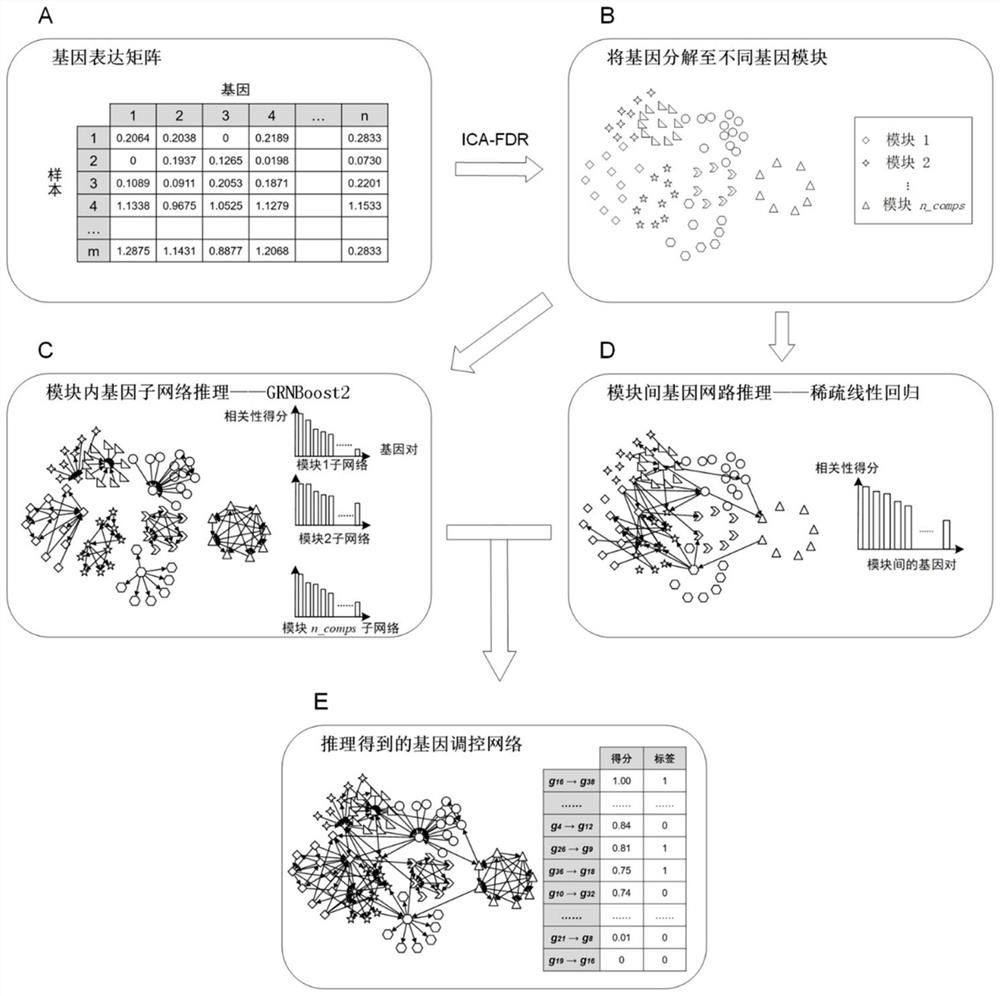
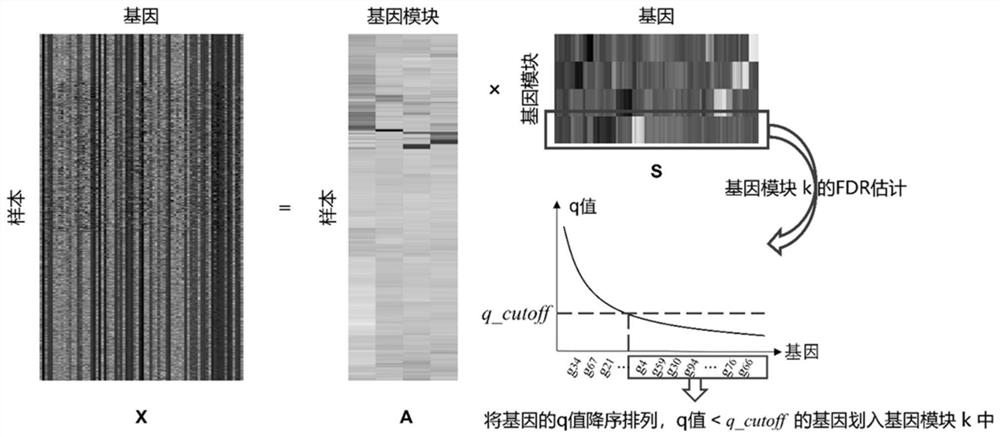
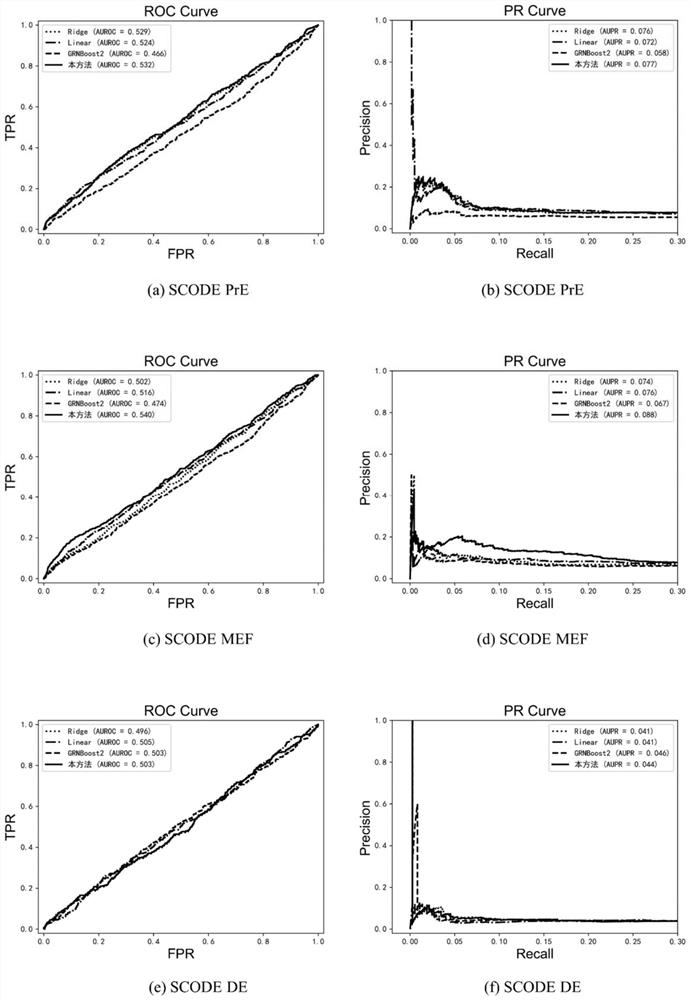
A Gene Network Inference Method Based on Modular Recognition
ActiveCN113066522BIncrease biological interpretabilityHigh speedBiostatisticsSystems biologyGradient boostingVersus gene
The invention discloses a gene network reasoning method based on modular identification. The method takes the expression information of n genes as the training set, and each gene has m samples; uses the ICA-FDR algorithm to identify the gene modules and divides the n genes into different gene modules; the regulation relationship within the gene module The algorithm based on gradient boosting tree is used for inference, and the regulatory relationship between gene modules is inferred by the algorithm based on sparse regression, and the correlation score of each gene pair is obtained; for each gene module and inter-module reasoning The obtained correlation scores were respectively normalized and merged, and sorted in descending order to obtain the final gene regulatory network. The invention provides a seamless fusion framework for gene module identification and gene network reasoning, improves the accuracy of gene module identification, and increases the interpretability of gene regulation network functions.
Owner:ZHEJIANG UNIV
A method for improving the fermentative production of glucaric acid by Saccharomyces cerevisiae engineering strains
The invention discloses a method for producing glucaric acid by improving saccharomyces cerevisiae engineered strain fermentation, and belongs to the technical field of bioengineering. According to the method disclosed by the invention, Delta sites of multiple copies in a saccharomyces cerevisiae genome are used as exogenous gene module integration sites, so that one-time integration of an exogenous gene module is realized so as to fulfil the aim of expression of the multiple copies. According to the method disclosed by the invention, through an integration expression way, gene engineering bacteria subjected to the integration of genome Delta sites is passaged for 20 times; and under the same fermentation condition, the yield of the glucaric acid is still 3.8g / L, and the method has an important application meaning on industrial production of the glucaric acid.
Owner:JIANGNAN UNIV
cerna competition module identification method, device, electronic equipment and storage medium
ActiveCN111383709BAccurate identificationCharacter and pattern recognitionProteomicsEngineeringMirna target gene
The invention provides a ceRNA competition module identification method, device, electronic equipment and storage medium, and relates to the field of gene identification. The ceRNA competition module recognition method includes: respectively according to the RNA 1 expression matrix and RNA 2 expression matrix to obtain RNA 1 Gene modules and RNA 2 gene module, where RNA 1 and RNA 2 are miRNA target genes. Based on a priori miRNA-target gene regulatory relationship data, miRNA expression matrix, RNA 1 Gene modules and RNA 2 The gene module identifies a ceRNA competition module that satisfies the conditions, wherein the ceRNA competition module includes an internal competition module and an external competition module. Due to the consideration of ceRNA internal and external competition, the strength of ceRNA internal competition and external competition was measured at the module level, as well as the influence of miRNAs on ceRNA internal and external competition. Therefore, the protocol provided by the present invention can accurately identify ceRNA competition modules.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA +1
System for detecting hypertension-related genes and analyzing gene functions
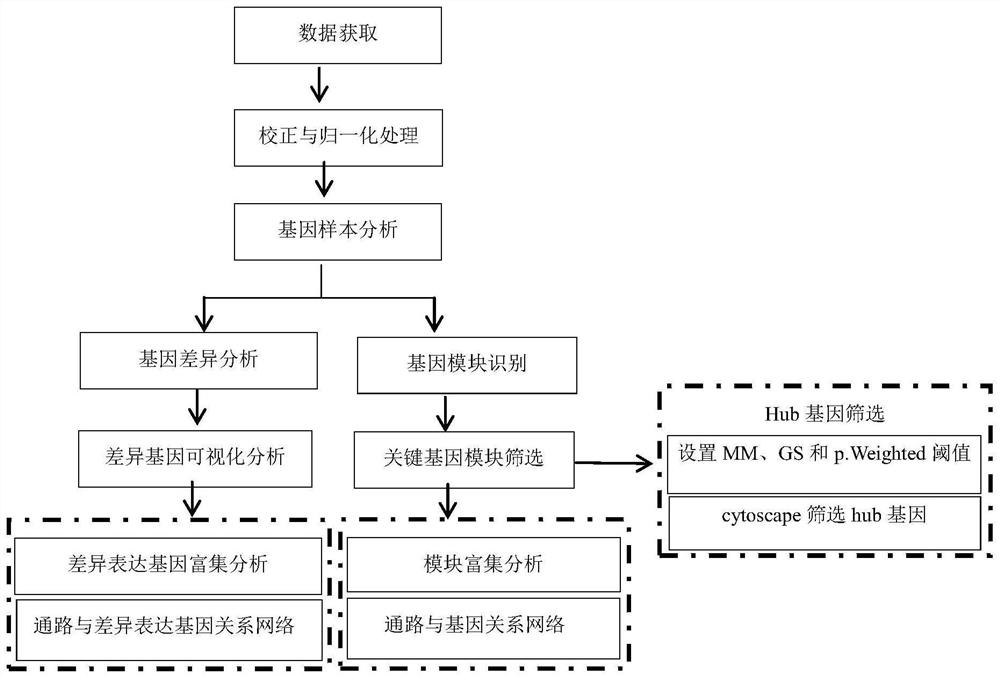
PendingCN114566222AReduce research costsEasy to implementData visualisationBiostatisticsSynexpressionDifferentially expressed genes
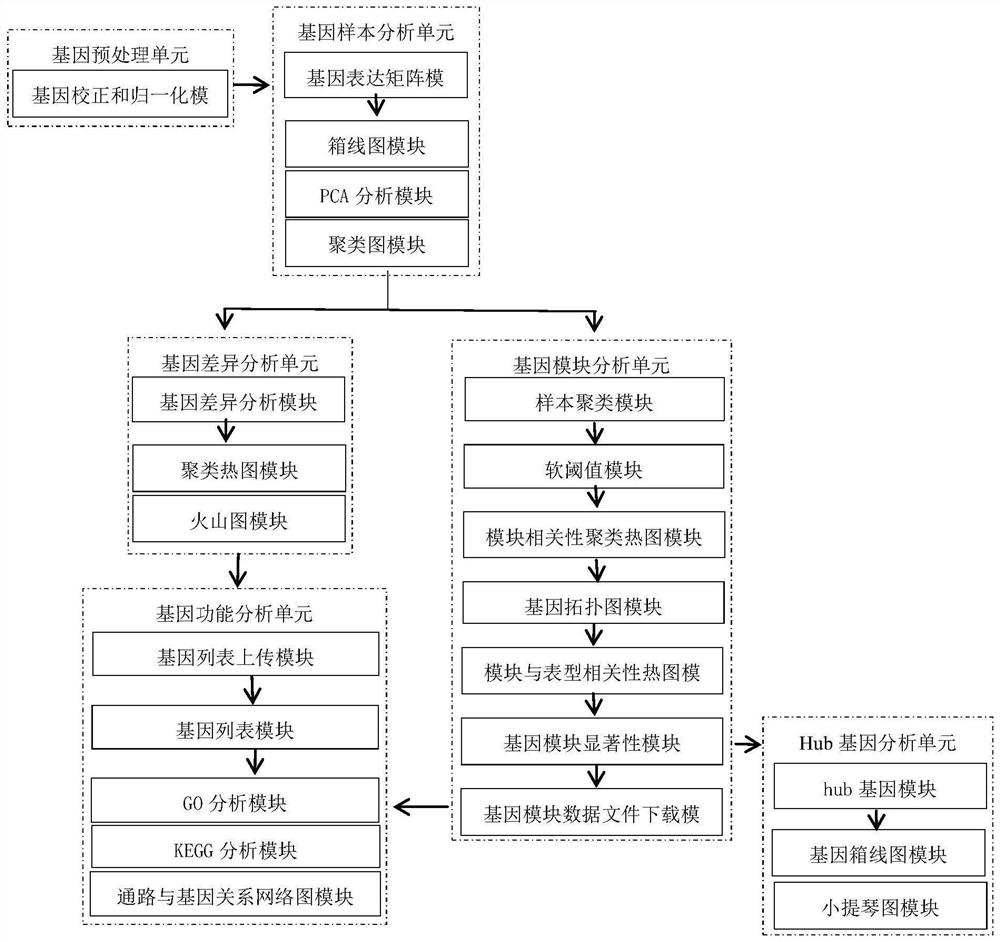
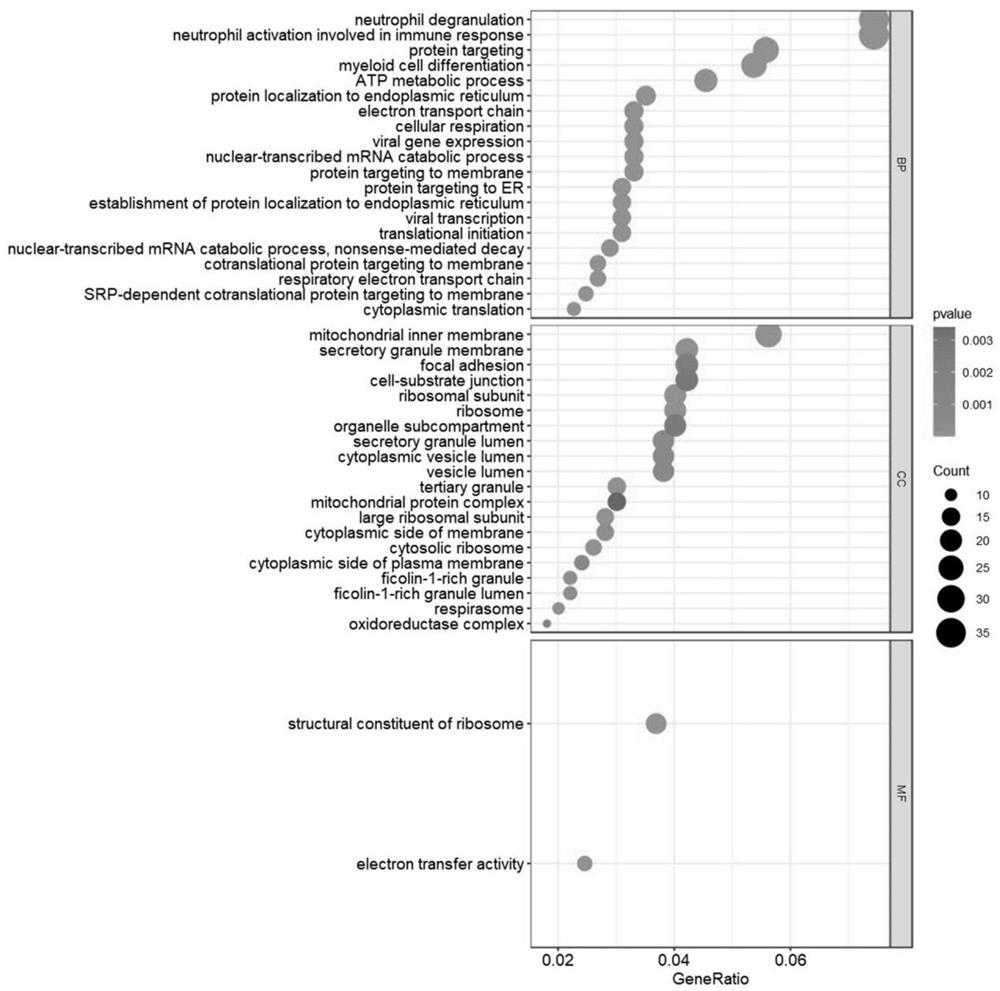
The invention discloses a system for detecting hypertension related genes and analyzing gene functions. Comprising a gene preprocessing unit used for preprocessing hypertension gene expression data to obtain a gene expression matrix; the gene sample analysis unit is used for analyzing a sample distribution condition based on the gene expression matrix; the gene difference analysis unit is used for screening differential genes with significant changes on the basis of the gene expression matrix to obtain a gene difference matrix; the gene module analysis unit is used for determining gene modules and screening key modules through a weighted gene co-expression network method based on the gene expression matrix; the Hub gene analysis unit is used for screening Hub genes in the key genes; and the gene function analysis unit is used for carrying out function enrichment analysis and network analysis of a pathway and gene relationship on the differentially expressed gene or the gene module. The method can assist scientific researchers who are not familiar with hypertension gene related databases and analysis processes in accurately and effectively finding genes and pathways related to complex diseases such as hypertension.
Owner:QINGHAI NORMAL UNIV
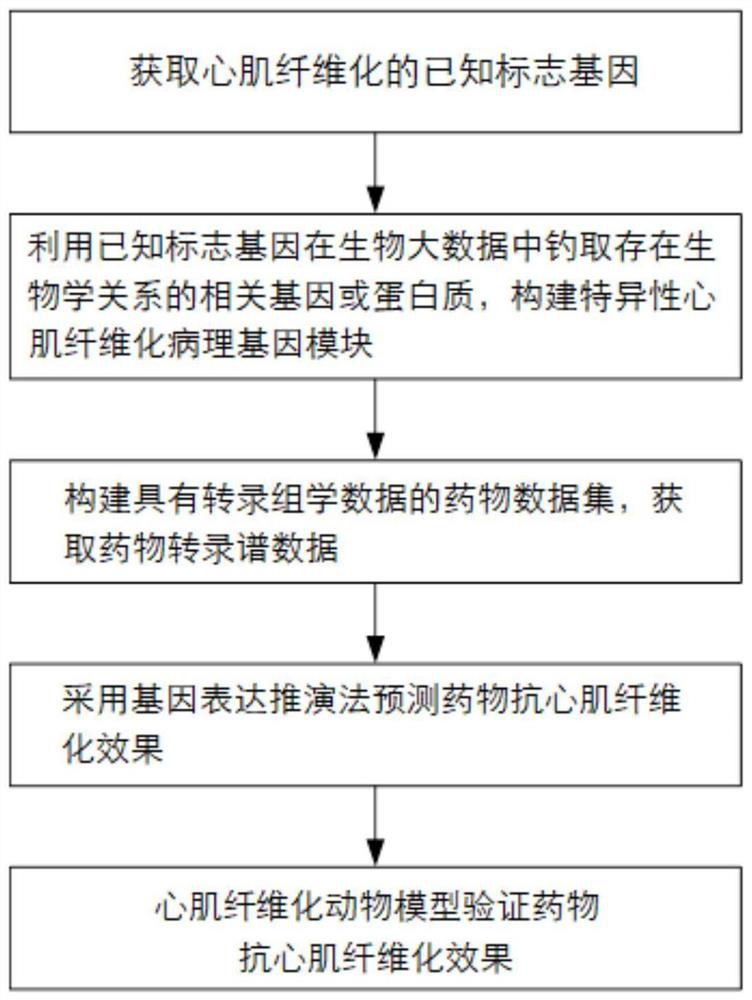
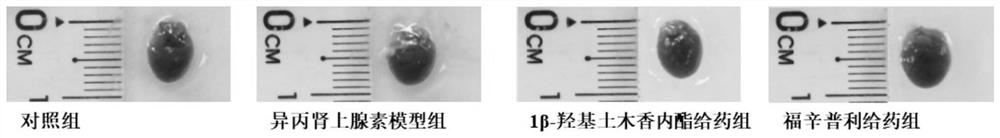
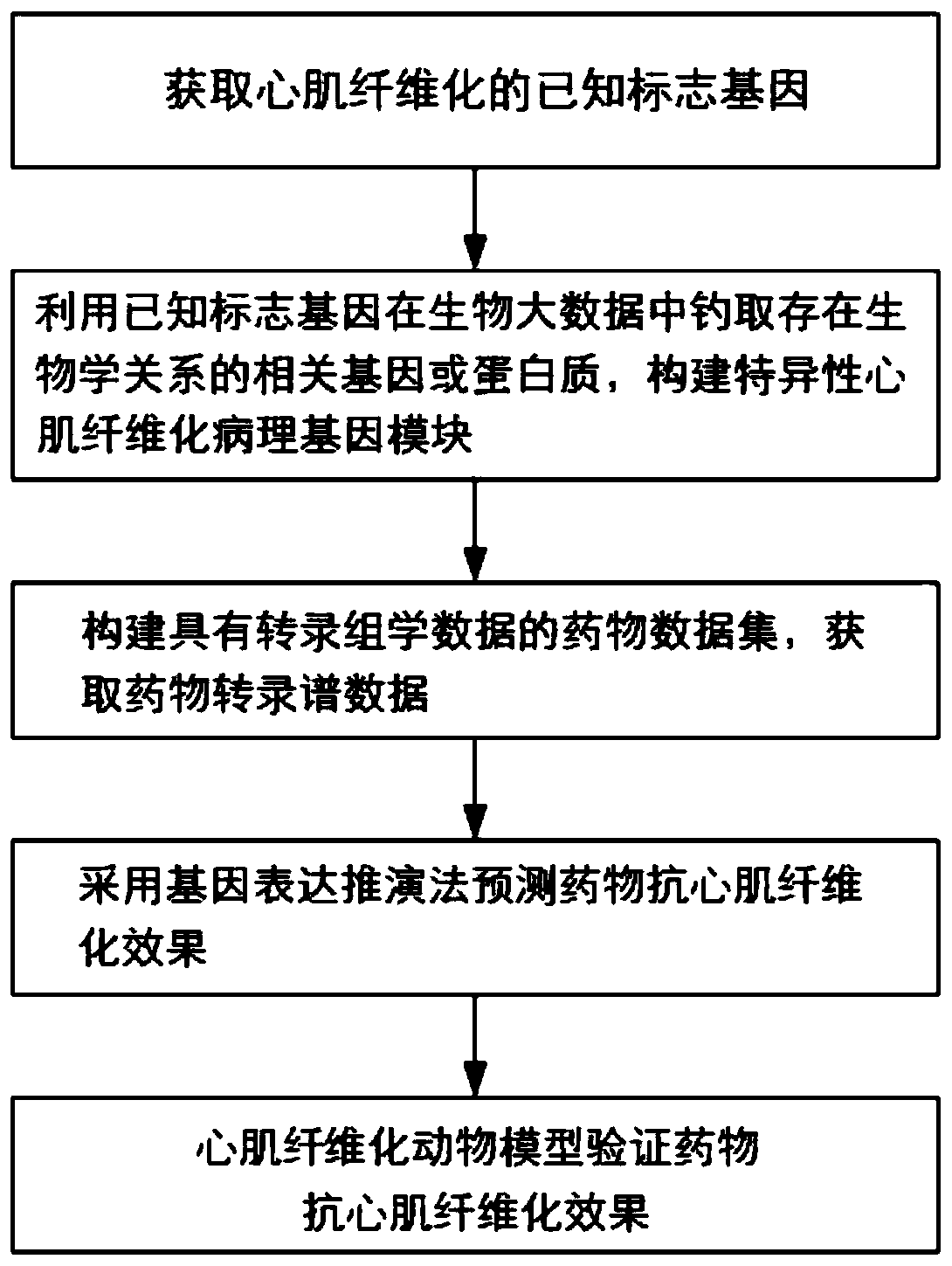
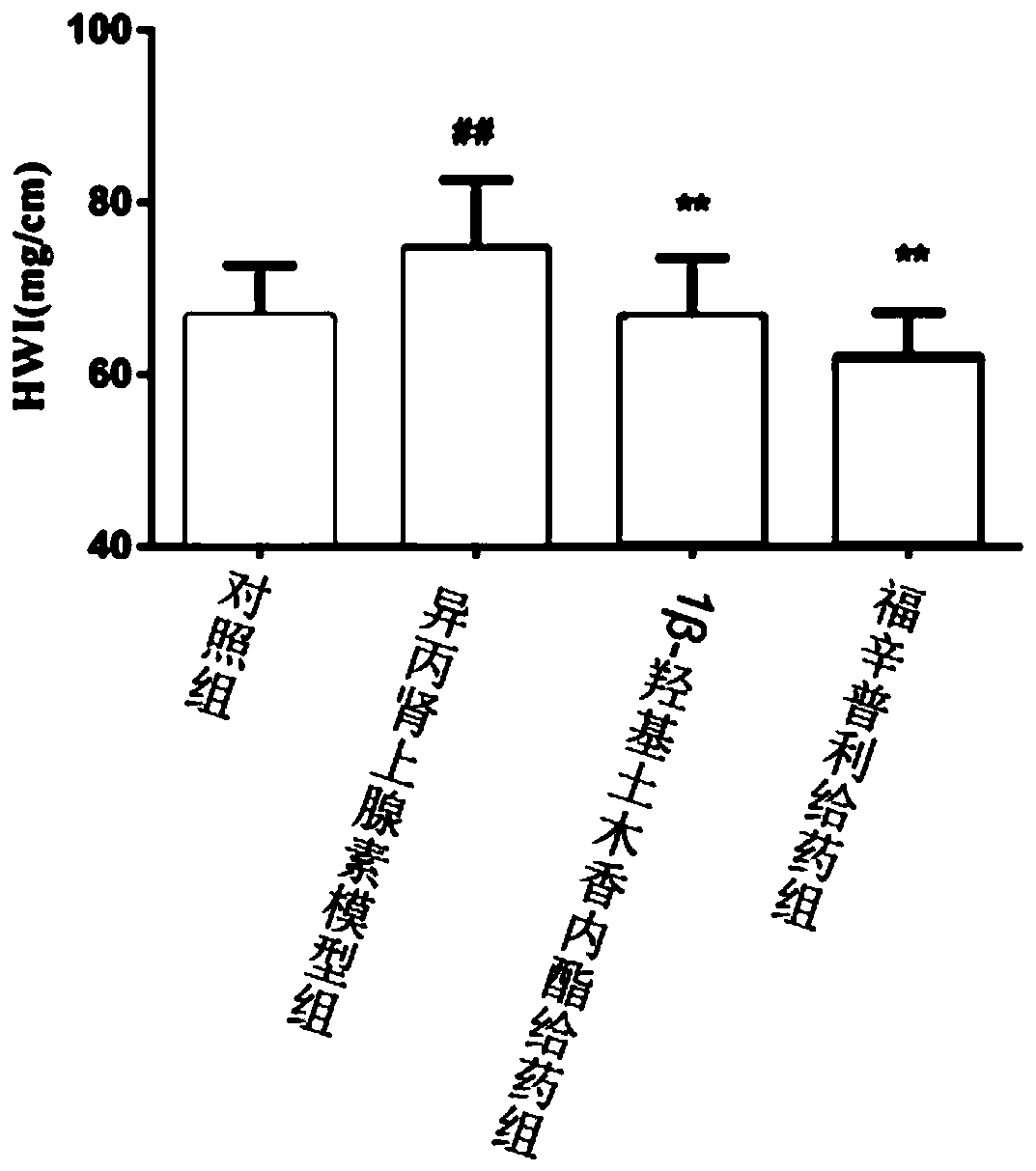
Screening method for anti-myocardial fibrosis drugs
The invention discloses a method for screening anti-myocardial fibrosis drugs, specifically: obtaining known marker genes of myocardial fibrosis; using the known marker genes to catch biological relationship with the known marker genes in biological big data Other genes or proteins, and limited to the genes that can be expressed in cardiac tissue, construct a specific myocardial fibrosis pathological gene module; construct a drug data set with transcriptomics data, and obtain drug transcription profile data; use gene expression deduction method according to drug The transcriptional profile data of the corresponding drug predicts the anti-myocardial fibrosis ability of the corresponding drug, and screens the desired drug according to the anti-myocardial fibrosis ability of the drug. Based on the different interactions between myocardial fibrosis marker genes and other genes, the present invention constructs a tissue-specific myocardial fibrosis module, screens potential anti-myocardial fibrosis drugs by computer, screens in batches, improves accuracy, high efficiency, and low cost .
Owner:SHANXI AGRI UNIV
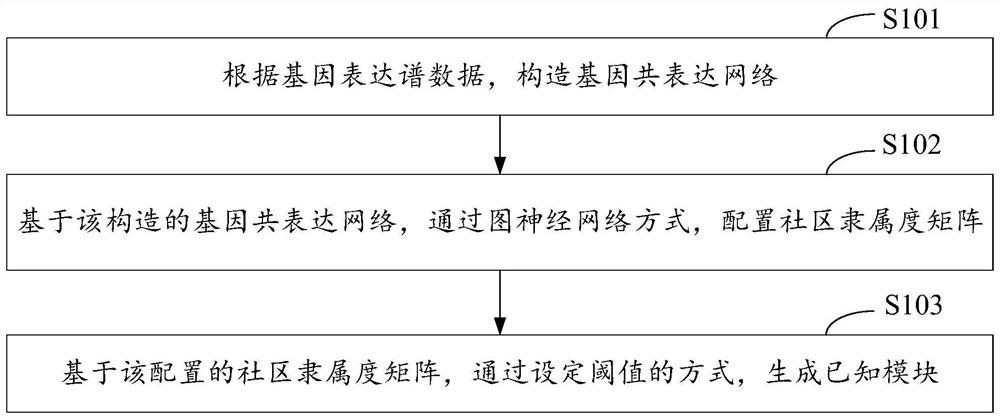
Gene module mining method, device, and computer equipment based on graph neural network
ActiveCN113611366BScale upImprove timing performanceBiostatisticsCharacter and pattern recognitionEngineeringGraph neural networks
The invention discloses a gene module mining method, device and computer equipment based on a graph neural network. Wherein, the method includes: constructing a gene co-expression network according to the gene expression profile data, and based on the constructed gene co-expression network, configuring a community membership matrix through a graph neural network, and a community membership matrix based on the configuration , generate known modules by setting thresholds. Through the above method, it is possible to configure the community affiliation matrix through graph neural network representation learning, and then generate known modules by setting thresholds, so that multiple genes with dense connections can be assigned to the gene module mining results. different modules.
Owner:HARBIN INST OF TECH SHENZHEN GRADUATE SCHOOL
Molecular classification and application of multiple myeloma
The invention discloses a multiple myeloma molecular subtype and application. The product provided by the invention comprises a substance for obtaining or detecting 97 gene expressions in a multiple myeloma cancer patient to be detected. The product further comprises equipment for operating a multiple myeloma Bayes classifier. In the invention, a gene module (MCL1-M for short) co-expressed with anMCL1 gene is identified, and the gene module is applied to divide multiple myeloma into two main subtypes, namely an MCL-M-High subtype and an MCL-M-Low subtype. The two subtypes have prognosis and genetics characteristics which are significantly different.
Owner:北京瑞牧西康医疗器械有限公司
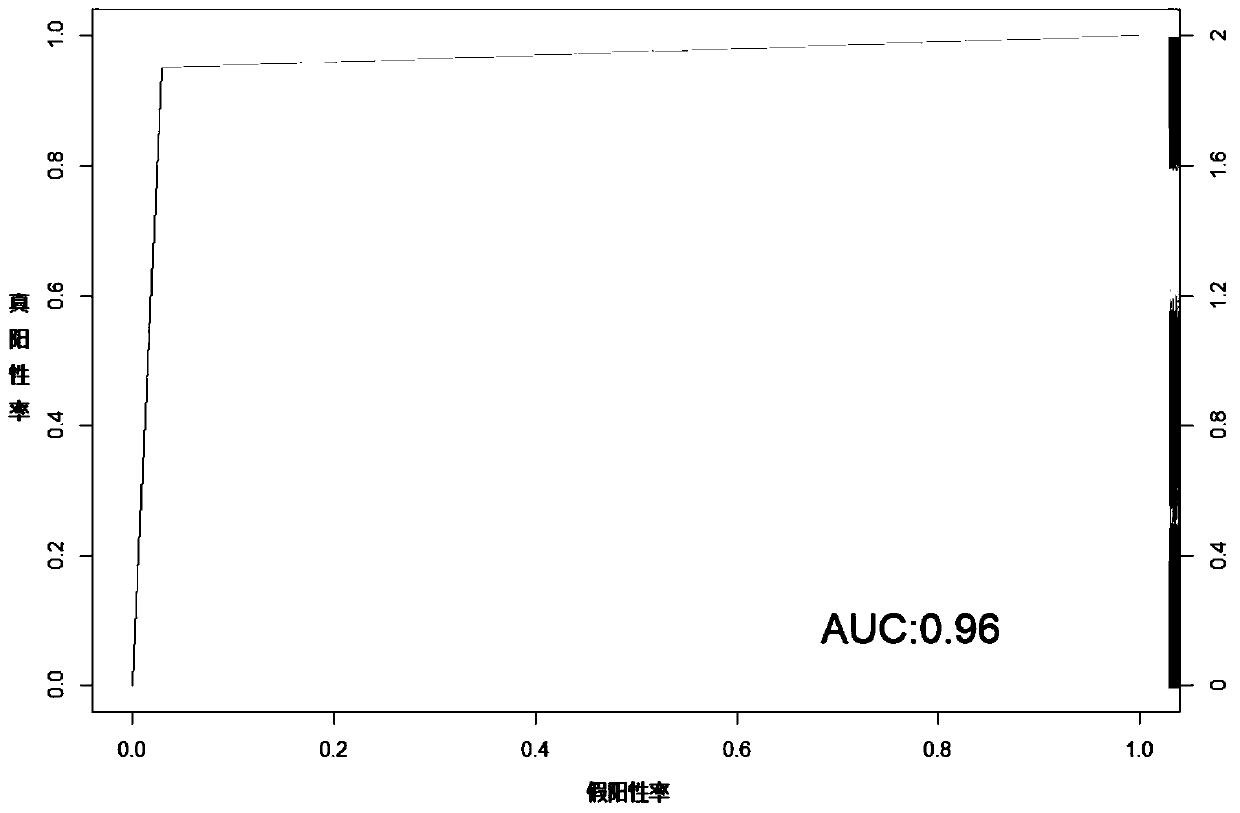
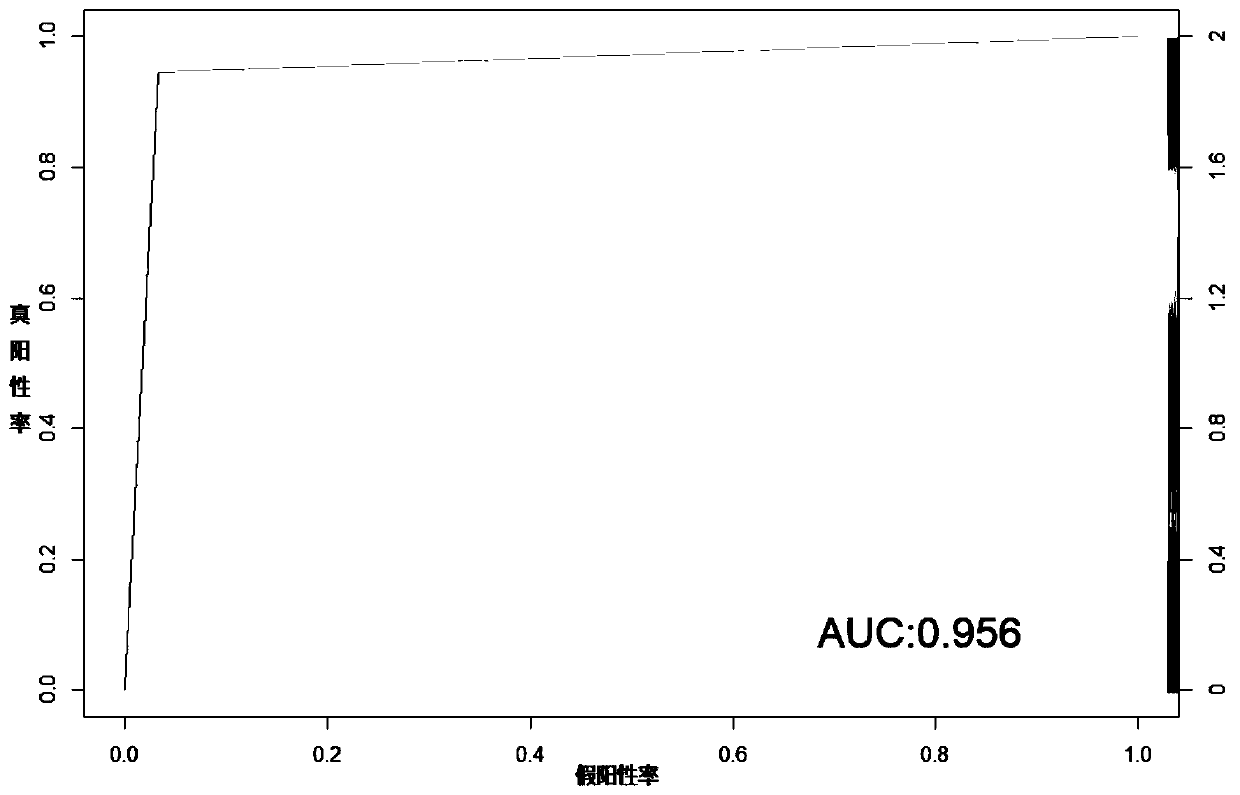
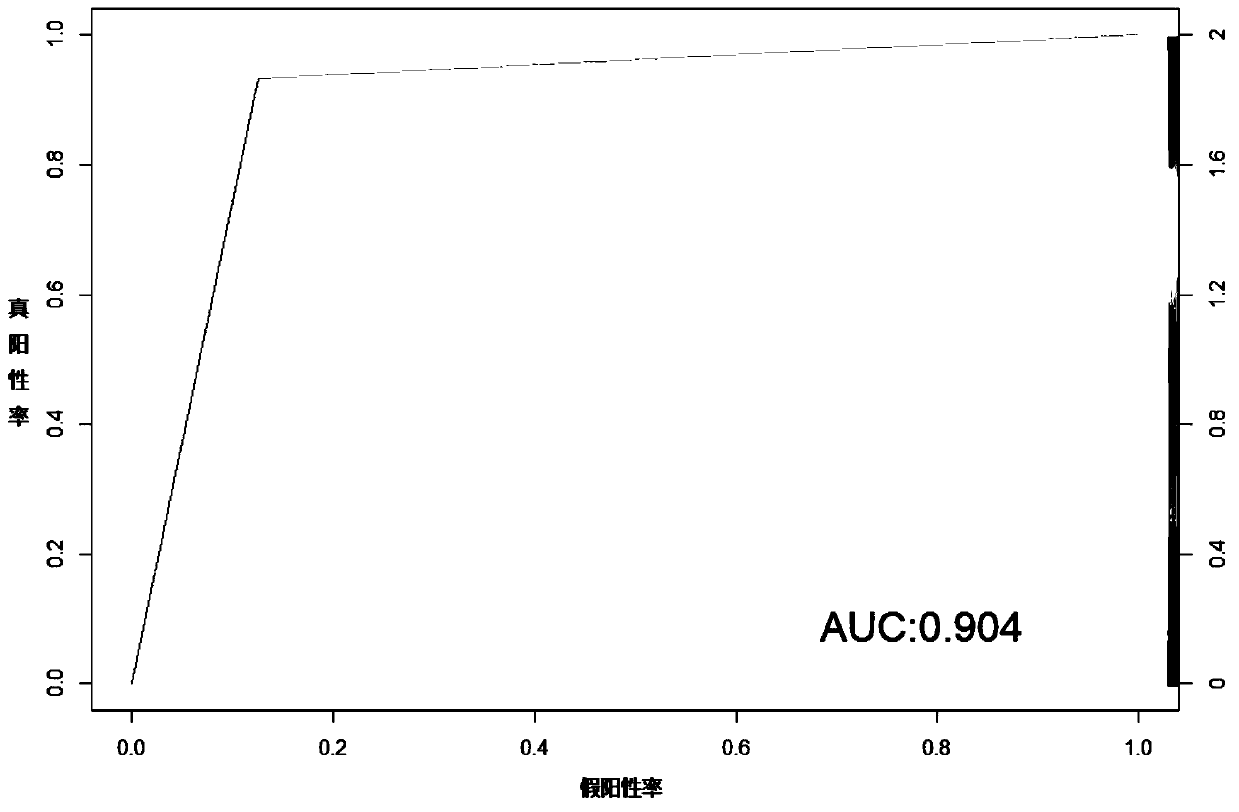
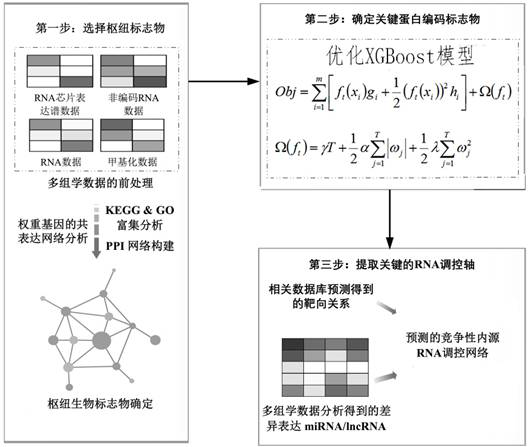
Method and system for multidimensional target prediction of disease-associated non-coding RNA regulatory axis
ActiveCN113921084BImprove reliabilityBiostatisticsInstrumentsSynexpressionDifferentially expressed genes
The invention provides a method and system for multi-dimensional targeting prediction of disease-related non-coding RNA regulatory axis, which belongs to the technical field of biological information processing based on machine learning, and uses multi-dimensional omics data to screen out the differentially expressed genes and co-expressed genes between the disease group and the control group. Express gene modules and perform enrichment analysis; based on the constructed protein-protein interaction network, process the screened differentially expressed genes and co-expressed genes to determine the composition of hub genes; among the identified hub genes, obtain key proteins Coding markers: using the constructed competitive endogenous RNA network, extract the non-coding RNA regulatory axis network including the key protein coding markers. The present invention can effectively predict complex disease-related endogenous competitive non-coding RNA regulatory network, and identify the key lncRNA-miRNA-mRNA regulatory axis, which helps to provide more promising candidates for the study of molecular pathogenic mechanisms of complex diseases Or, provide potential molecular markers for the development of precision medicine.
Owner:SHANDONG UNIV QILU HOSPITAL
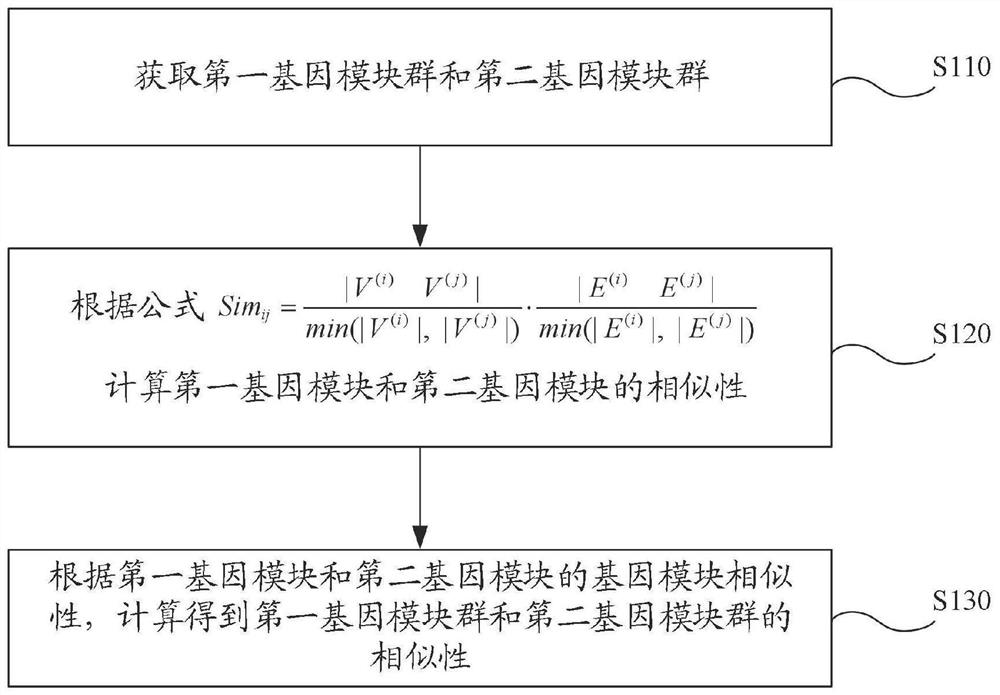
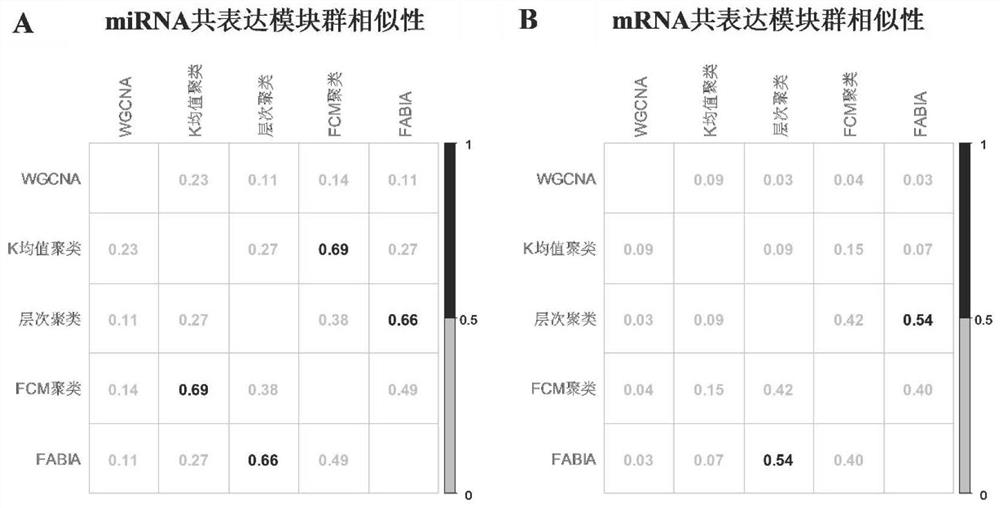
Similarity measurement method and device of gene module group, electronic equipment and storage medium
ActiveCN113947149AAccurate similaritySimilarity measureCharacter and pattern recognitionHybridisationGene recognitionComputer science
The invention provides a similarity measurement method and a device of a gene module group, electronic equipment and a storage medium, and relates to the technical field of gene recognition. The similarity measurement method for the gene module groups comprises the following steps: collecting a first gene module group and a second gene module group; calculating the similarity between the first gene module and the second gene module according to a formula; and calculating the similarity of the first gene module group and the second gene module group according to the gene module similarity of the first gene module and the second gene module. According to the method, based on a given formula, when the similarity between the first gene module group and the second gene module group is calculated, the gene interaction relation is considered, and the similarity between different gene module groups can be measured more accurately.
Owner:DALI UNIV
Screening method of anti-myocardial fibrosis drug
The invention discloses a screening method of an anti-myocardial fibrosis drug. The screening method specifically comprises the following steps: obtaining a known marker gene of myocardial fibrosis; fishing other genes or proteins which have a biological relationship with the known marker gene from biological big data by utilizing the known marker gene, and limiting the range to the genes which can be expressed by heart tissues to construct a specific myocardial fibrosis pathological gene module; constructing a drug data set with transcriptomics data, and obtaining drug transcriptomics data; predicting the anti-myocardial fibrosis capability of the corresponding drug according to the transcriptional spectrum data of the drug by adopting a gene expression deduction method, and screening therequired drug according to the anti-myocardial fibrosis capability of the drug. On the basis of different interactions of the myocardial fibrosis marker gene and other genes, a tissue-specific myocardial fibrosis module is constructed, anti-myocardial fibrosis potential drugs are screened through a computer, the batch screening is conducted, the accuracy is improved, the efficiency is high, and the cost is low.
Owner:SHANXI AGRI UNIV
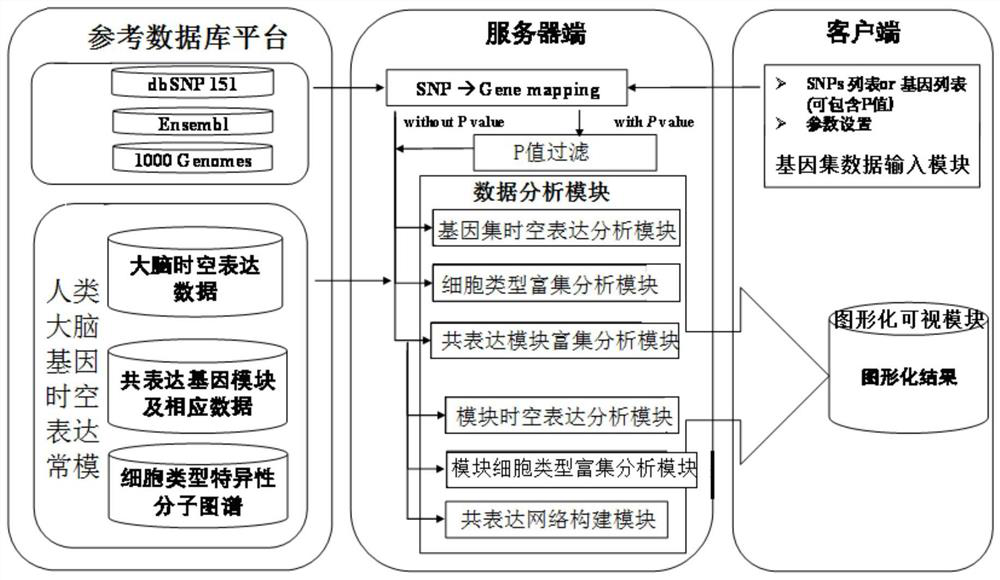
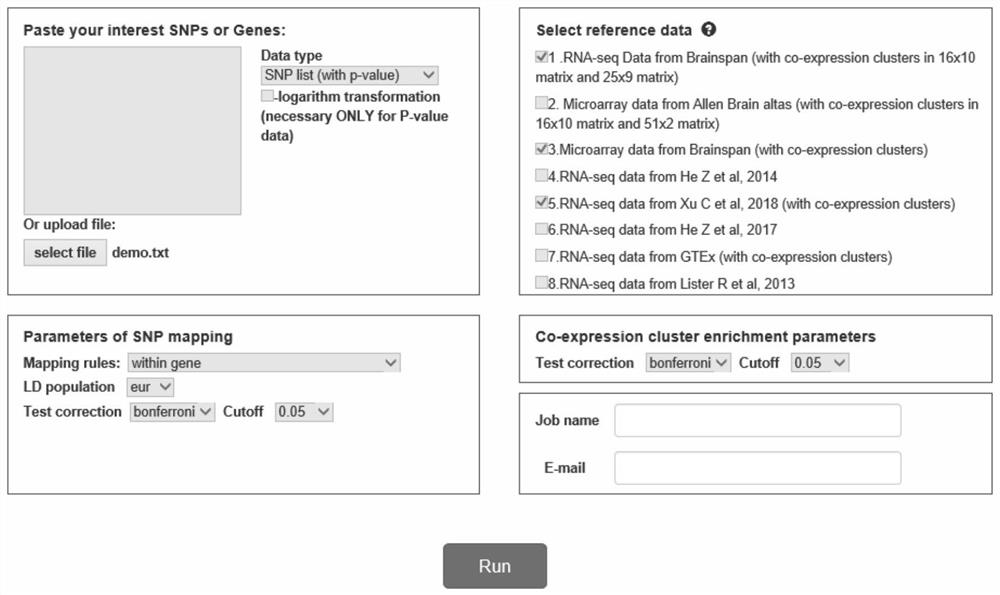
A human brain gene spatiotemporal expression pattern online analysis system and method thereof
ActiveCN110428867BEasy to understandRich correlationData visualisationBiostatisticsServer-sideGene sets
The invention discloses an online analysis system and method for gene human brain spatiotemporal expression patterns. The reference database platform includes human brain gene spatiotemporal expression norms, thousand genome database, Ensembl database and dbSNP database; human brain gene spatiotemporal expression norms Based on the constructed spatio-temporal framework of each brain region and each developmental stage, form the matrix expression pattern of genes or gene modules between each brain region and each developmental stage, as well as the enrichment matrix expression pattern between gene modules and cell types; There is a data analysis module on the server side; a gene set data input module and a graphic visualization module are set on the client side, the server side analyzes the spatio-temporal pattern of the input gene set data, and analyzes the genes through the graphic visualization module The results are presented graphically. The invention generates user-friendly and easy-to-understand visualization results on the client side, helps clarify the complex spatio-temporal expression patterns of genes, and reflects the relationship between the analyzed genes.
Owner:INST OF PSYCHOLOGY CHINESE ACADEMY OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com