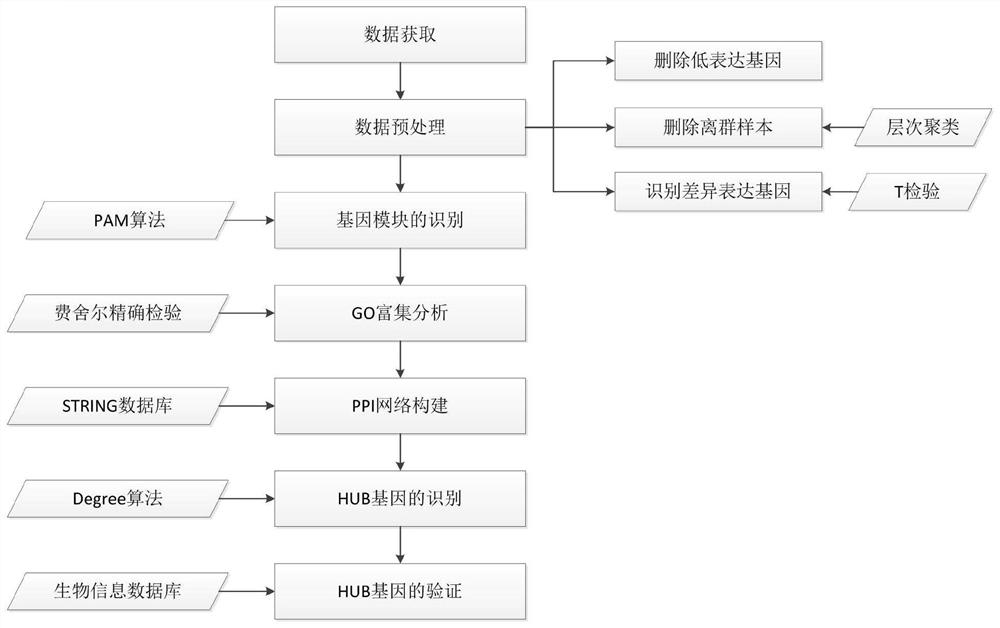
Gene expression data analysis method based on PAM clustering algorithm
A technology for gene expression and data analysis, applied in the field of data analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0081] Research on the mechanism of action of NSC319726 based on this method
[0082] (1) Preliminary identification results of genes
[0083] In this study, the T test was used to test the expression of each gene in the original data in the treatment group and the control group. After the conditional screening of P<=0.05, a total of 5044 genes with statistical significance were identified for further analysis.
[0084] (2) Use the PAM algorithm to mine functional gene modules
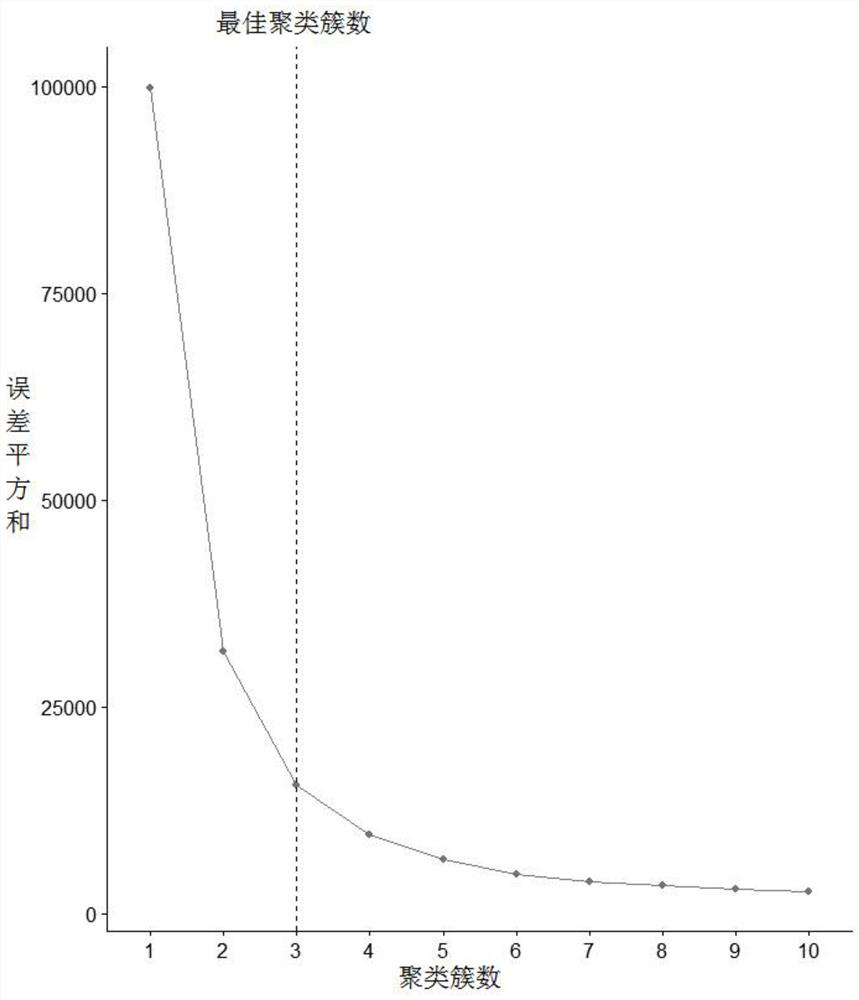
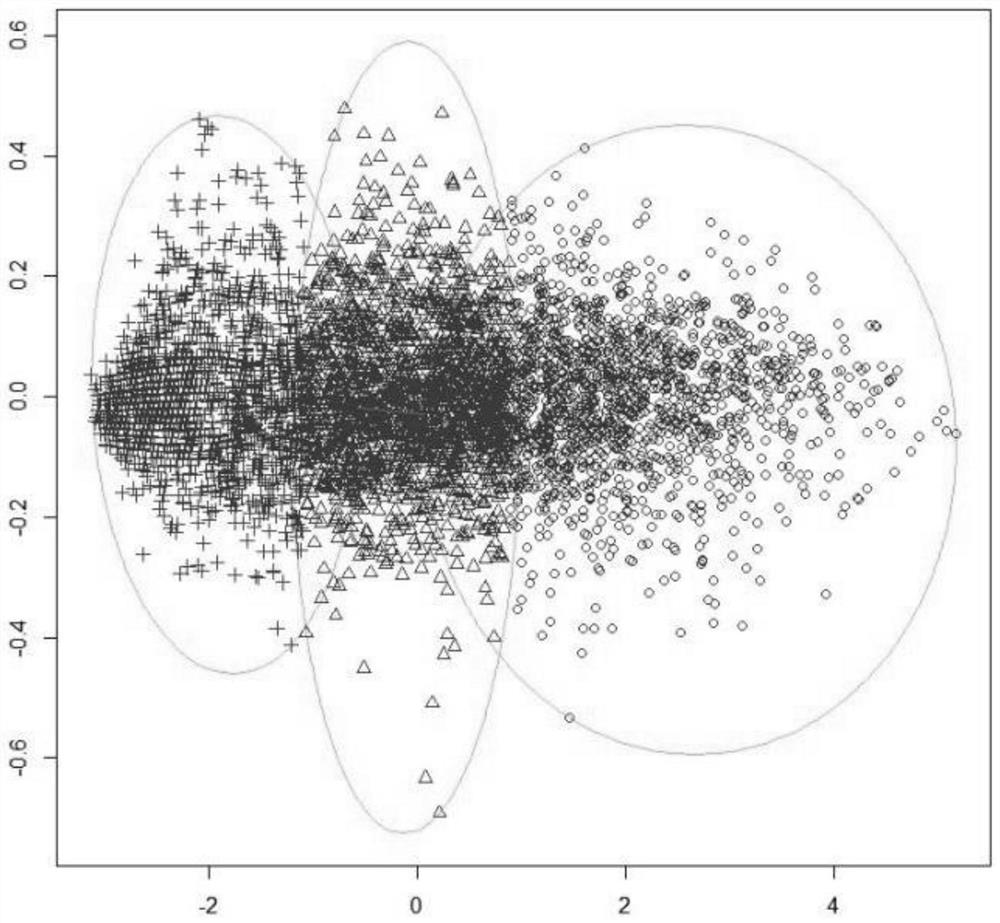
[0085] In this study, the elbow rule was used to further determine the number of clusters ( figure 2 ). Depend on figure 2 It can be seen that the optimal number of clusters in this study is 3. The expression of 5044 genes in the drug-administered group was clustered using the PAM algorithm, and the clustering results are shown in image 3 . Obviously, the 3 clusters obtained by the PAM algorithm are 3 gene modules, the module m1 contains 1599 genes, the module m2 contains 1964 genes, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com