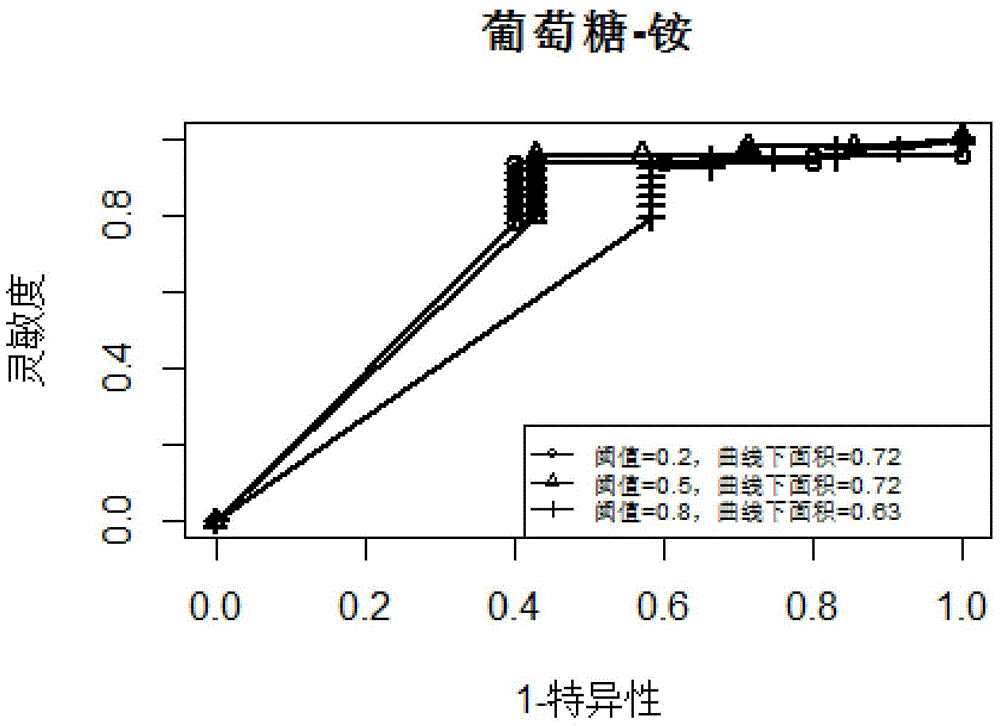
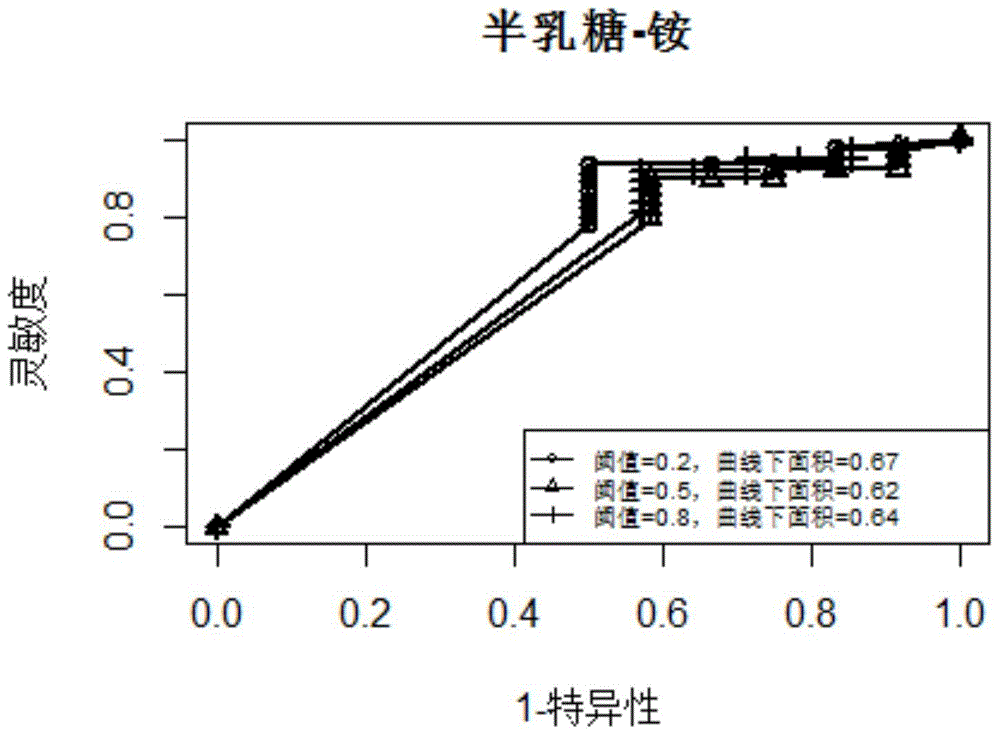
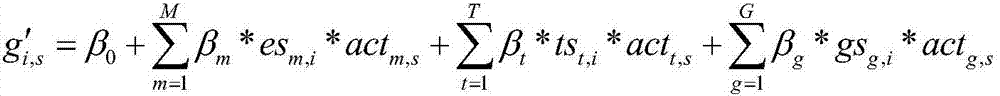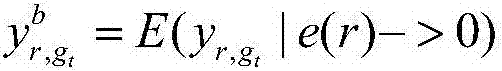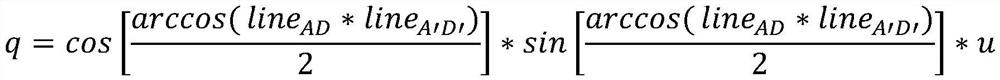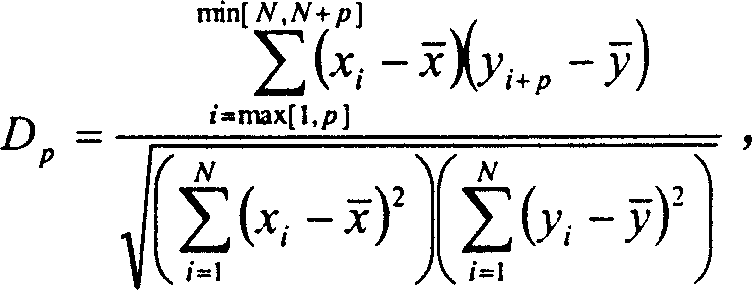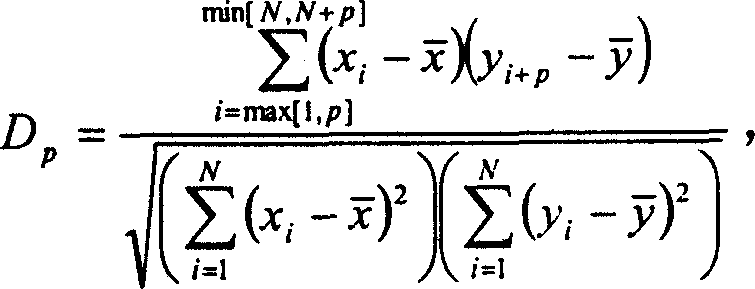Patents
Literature
105 results about "Gene regulatory network" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A gene (or genetic) regulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins. These play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).
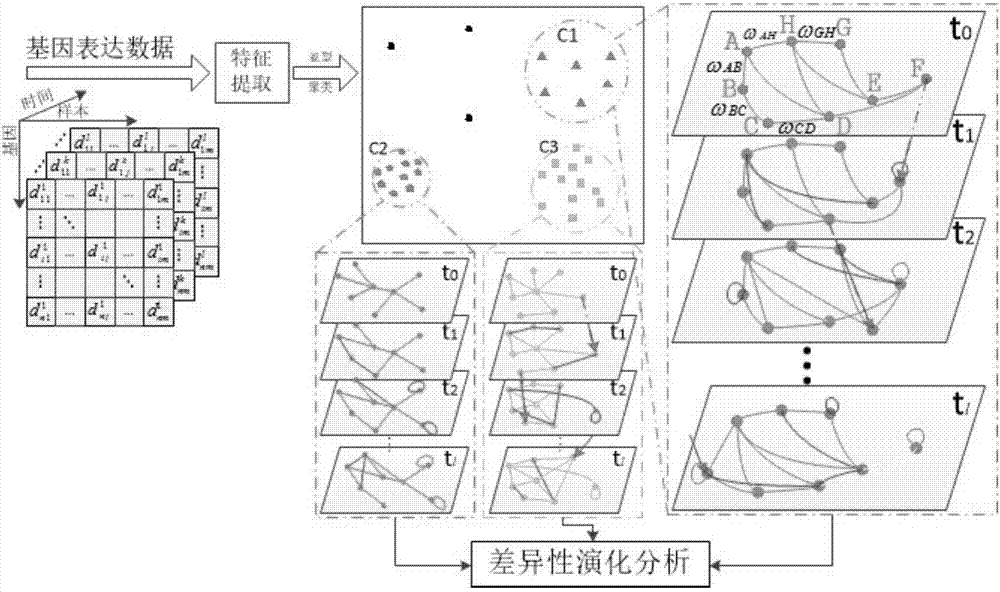
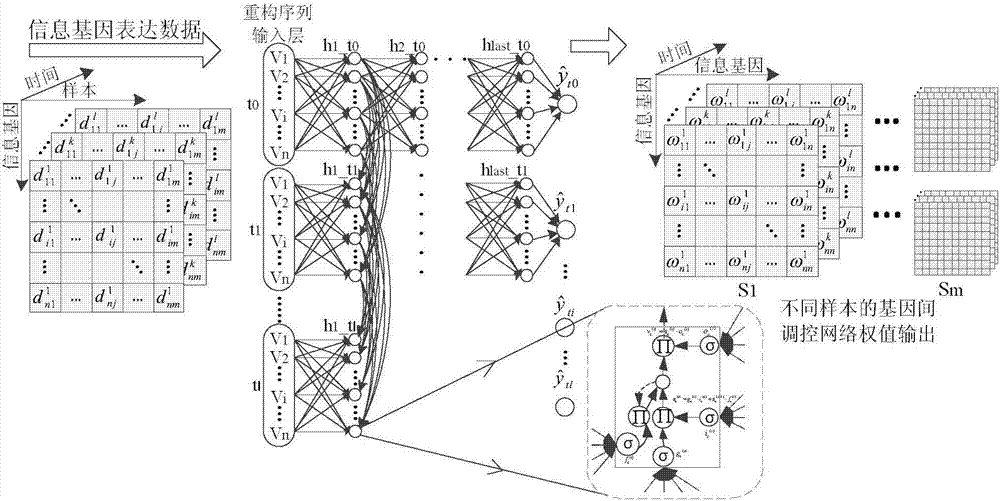
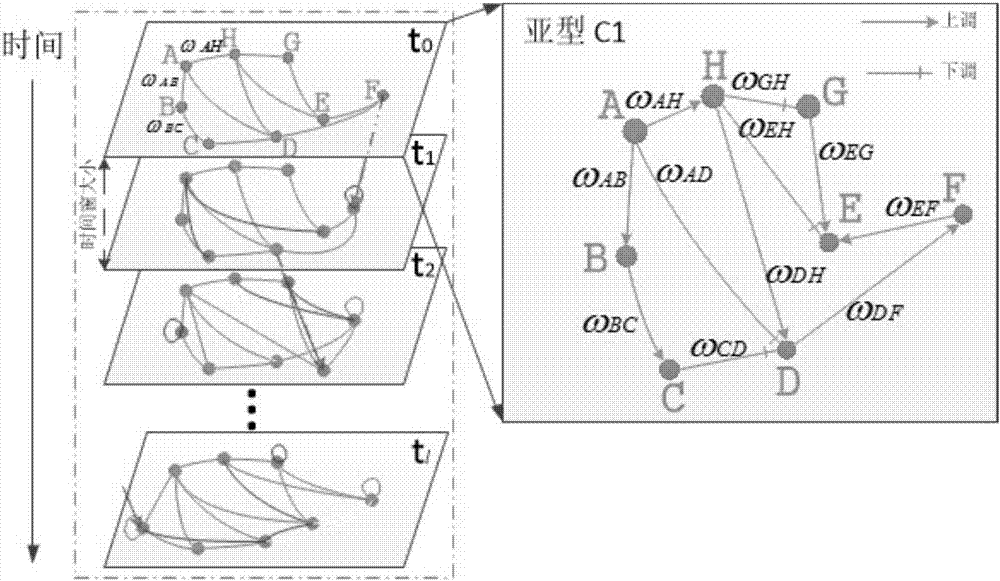
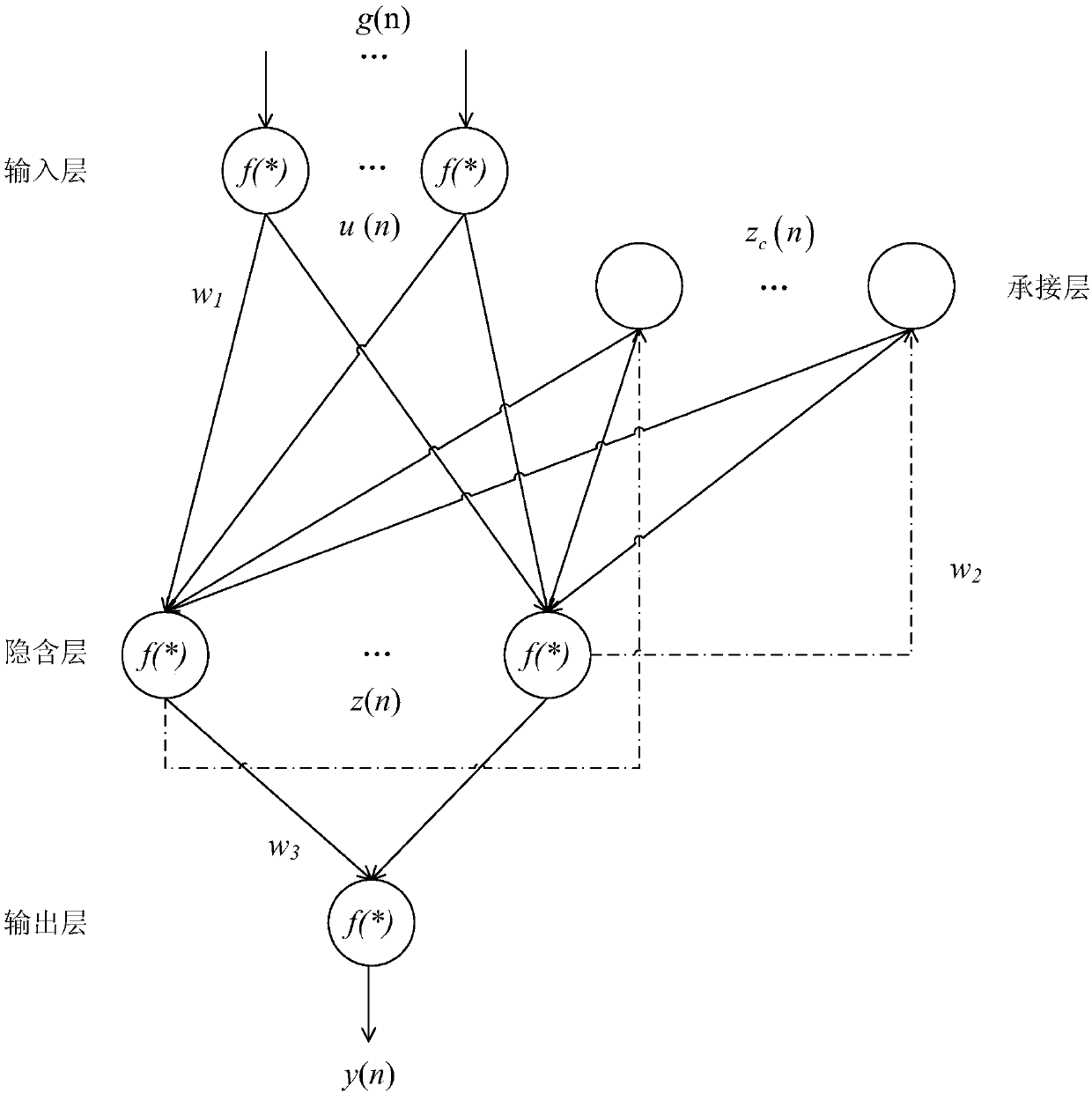
RNN-based gene regulatory network construction and dynamic differential analysis method
An RNN-based gene regulatory network construction and dynamic differential analysis method comprises the first step of gene dynamic regulation network construction based on a deepRNN, the second step of time-sequence variation and evolution analysis based on an intra-subtype dynamic regulation network, and the third step of network differential evolution analysis based on an inter-subtype dynamic regulation network, wherein the evolution analysis of different subtype networks comprises kinetic analysis, differential analysis and perturbation analysis. The RNN-based gene regulatory network construction and dynamic differentiation analysis method is high in precision.
Owner:ZHEJIANG UNIV OF TECH
Method for constructing genetic regulation network based on total-transcriptome high-throughput sequencing
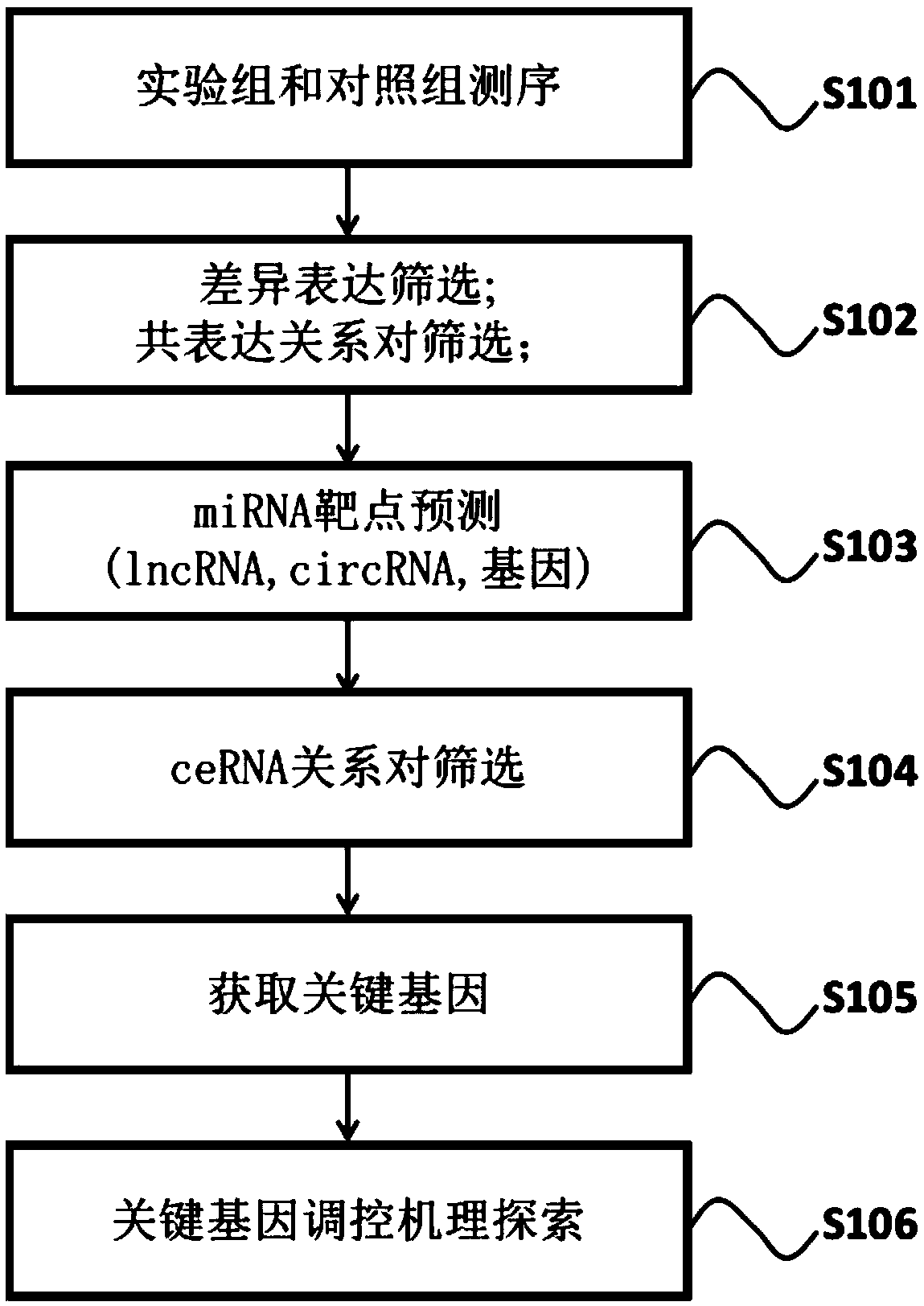
The invention relates to the technical field of biomedicine, and particularly relates to a method for constructing a genetic regulation network based on total-transcriptome high-throughput sequencing.The method comprises the steps that a disease related competitive endogenous RNA regulation network is constructed on the basis of total-transcriptome high-throughput sequencing data through the screening of differently expressed genes, lncRNA, circRNA and miRNA, the co-expression analysis of the genes, the lnc RNA and the circRNA and the miRNA combining target prediction, key genes in the regulation network are screened according to a random walking algorithm, the pathway in which the key genes are significantly enriched is determined through pathway enrichment analysis, and the biological mechanism of the key genes in the disease regulation process is analyzed combining with the genetic regulation relationship of the pathway. The invention provides methods for the verification of the disease related key genes and the construction of the regulation network of the key genes, and a novel approach is provided for the study of generating developing mechanisms of complicated diseases.
Owner:BIOMARKER TECH
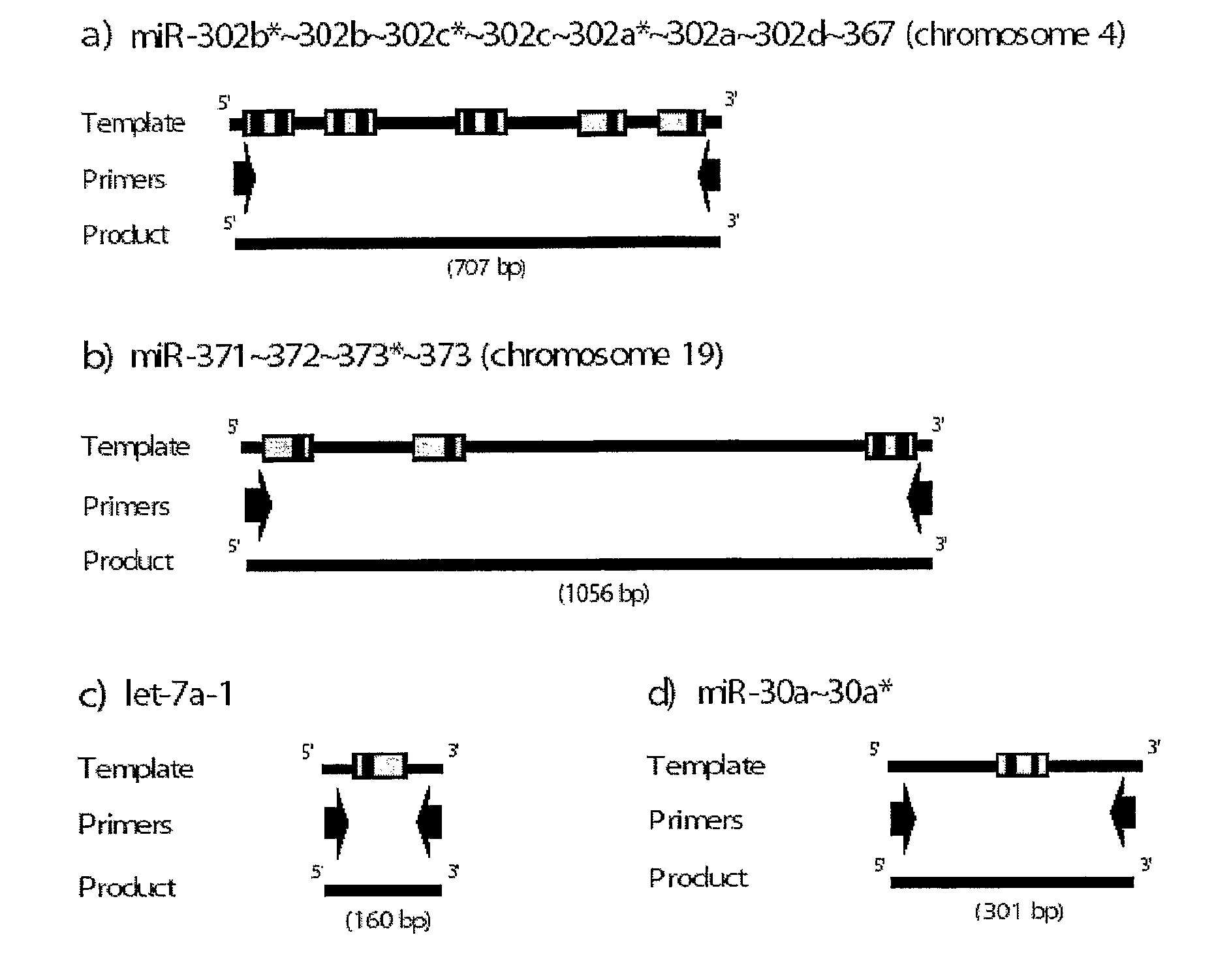
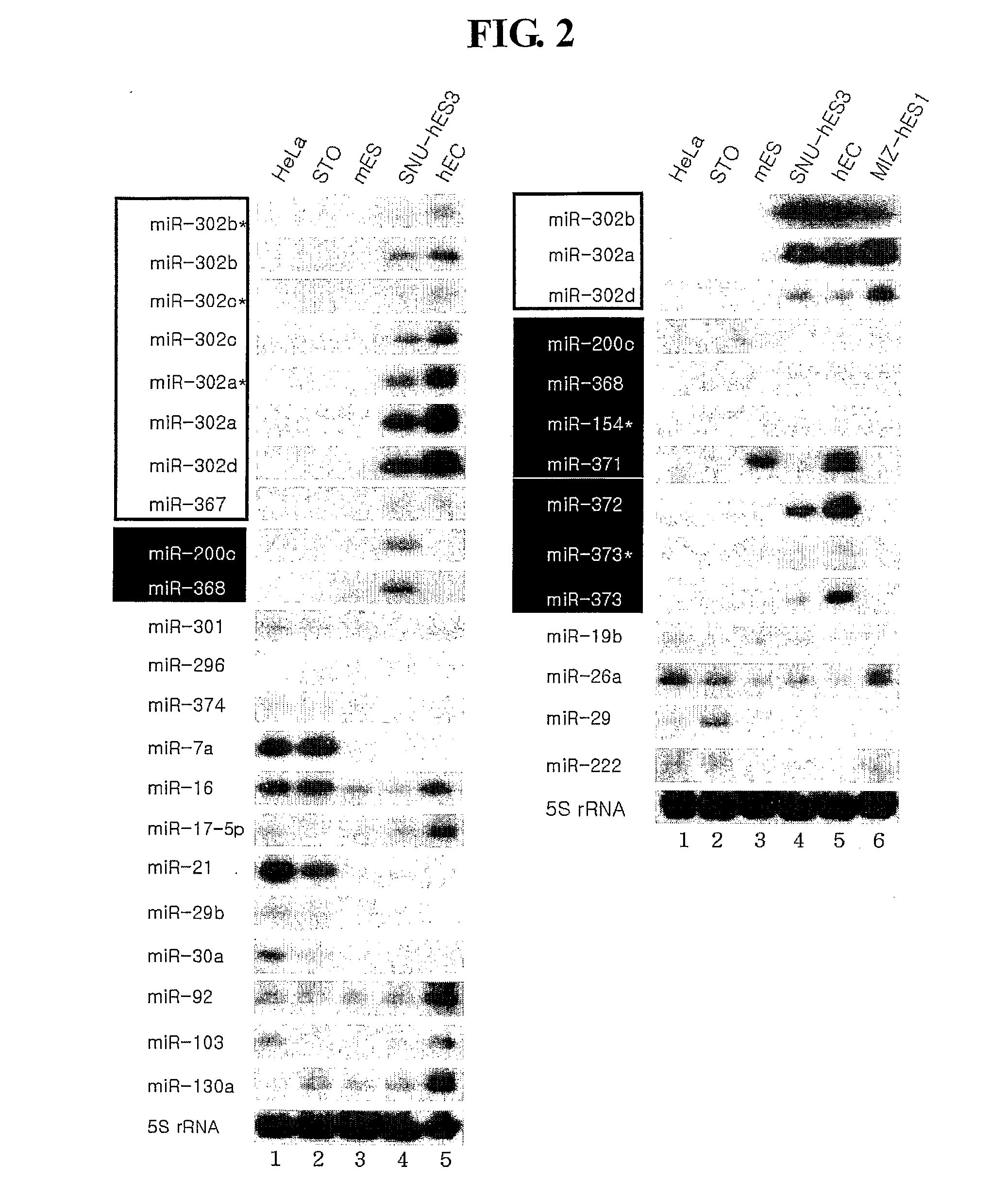
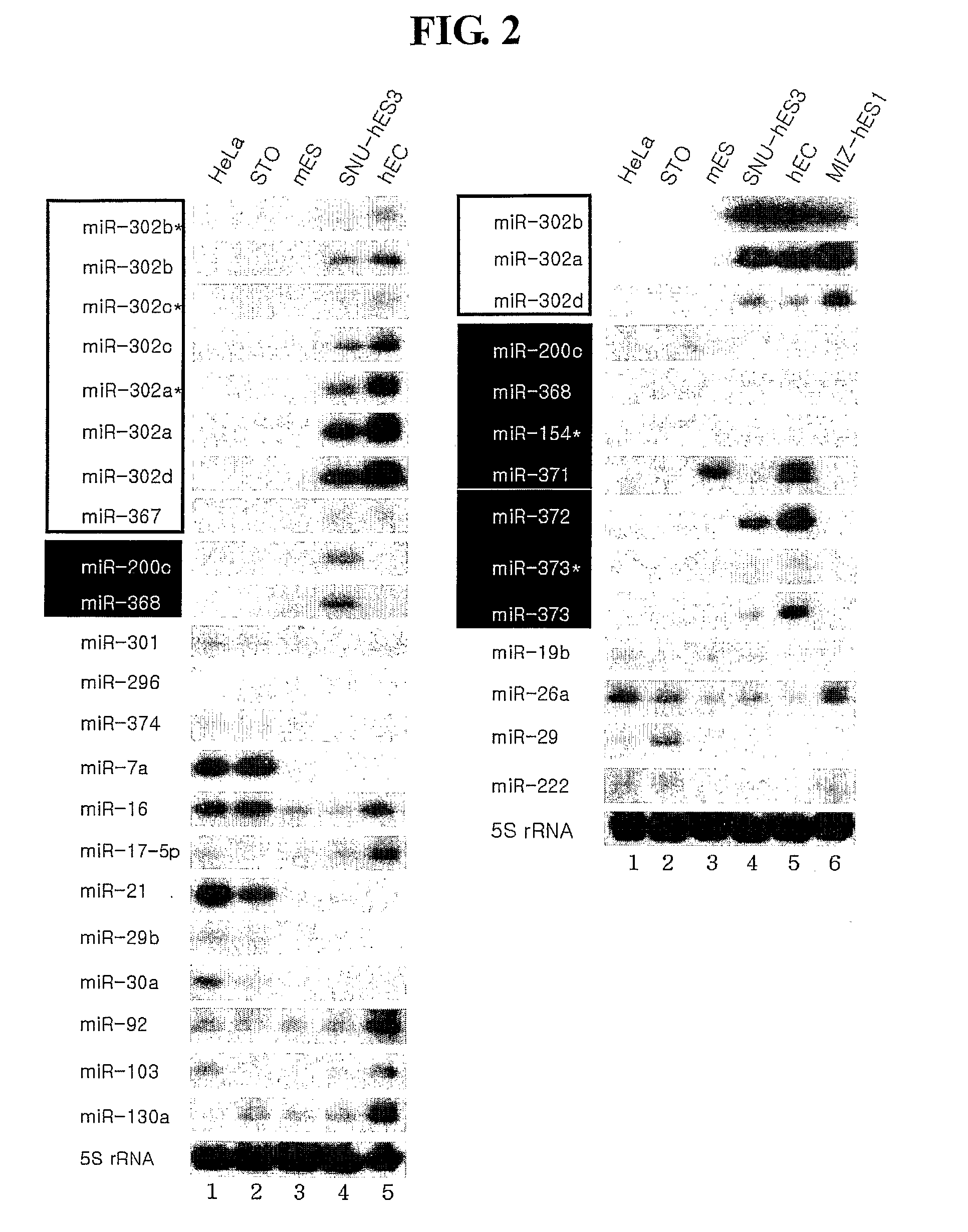
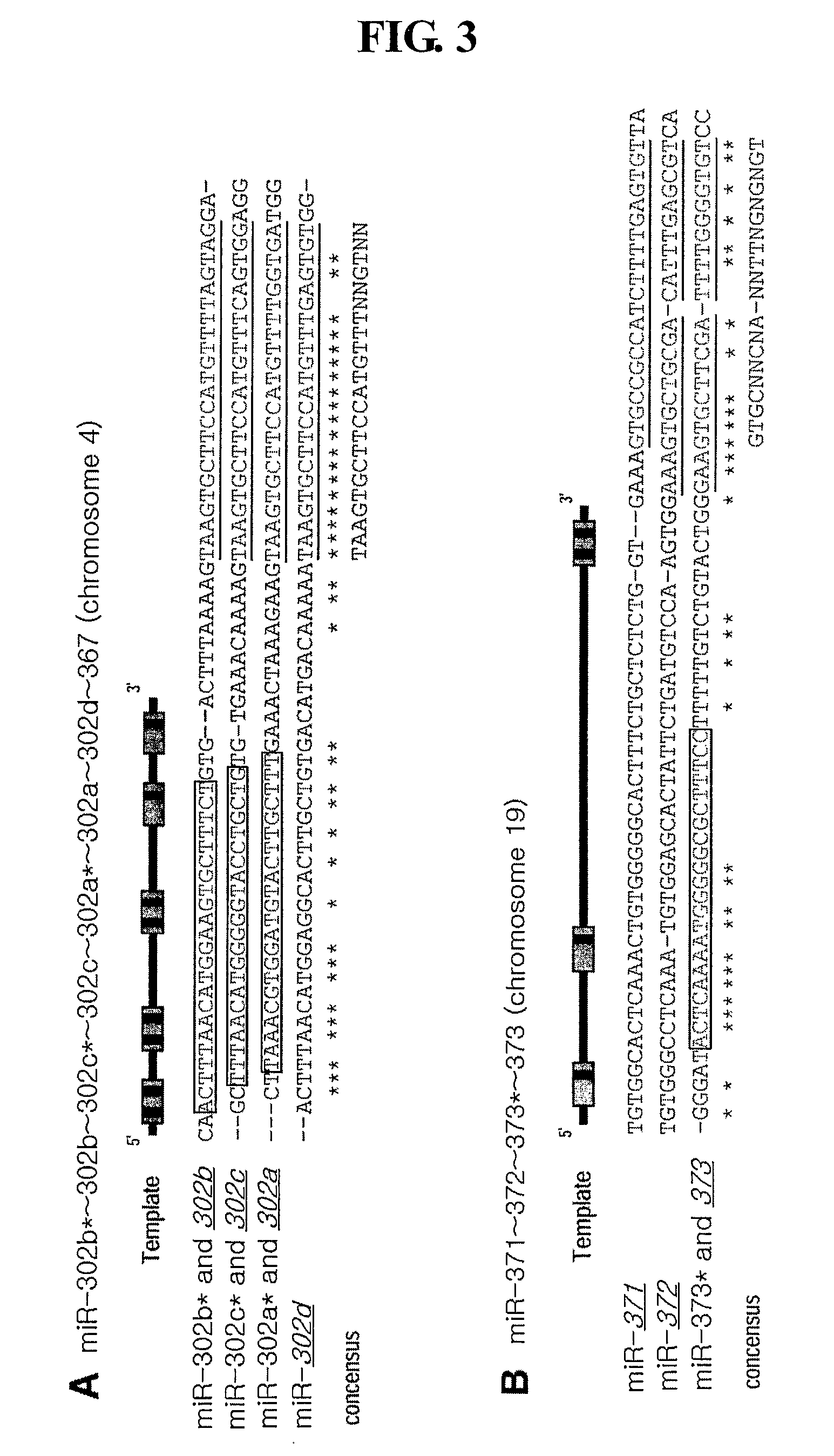
Novel Mirna Molecules Isolated from Human Embryonic Stem Cell
ActiveUS20080050722A1Good curative effectSugar derivativesMicrobiological testing/measurementDevelopmental stageStem Cell Isolation
The present invention relates to novel miRNA molecules, more particularly to novel miRNA molecules isolated from human embryonic stem cells. The miRNA molecules provided by the present invention can be usefully used as a molecular marker for early developmental stages of undifferentiated human embryonic stem cells. Also, the miRNA molecules of the present invention may play an important role in the regulation of mammalian embryonic stem cells. Therefore, the miRNA molecules can be usefully used for analyzing regulatory networks of human embryonic stem cells.
Owner:COLLEGE OF MEDICINE POCHON CHA UNIV IND
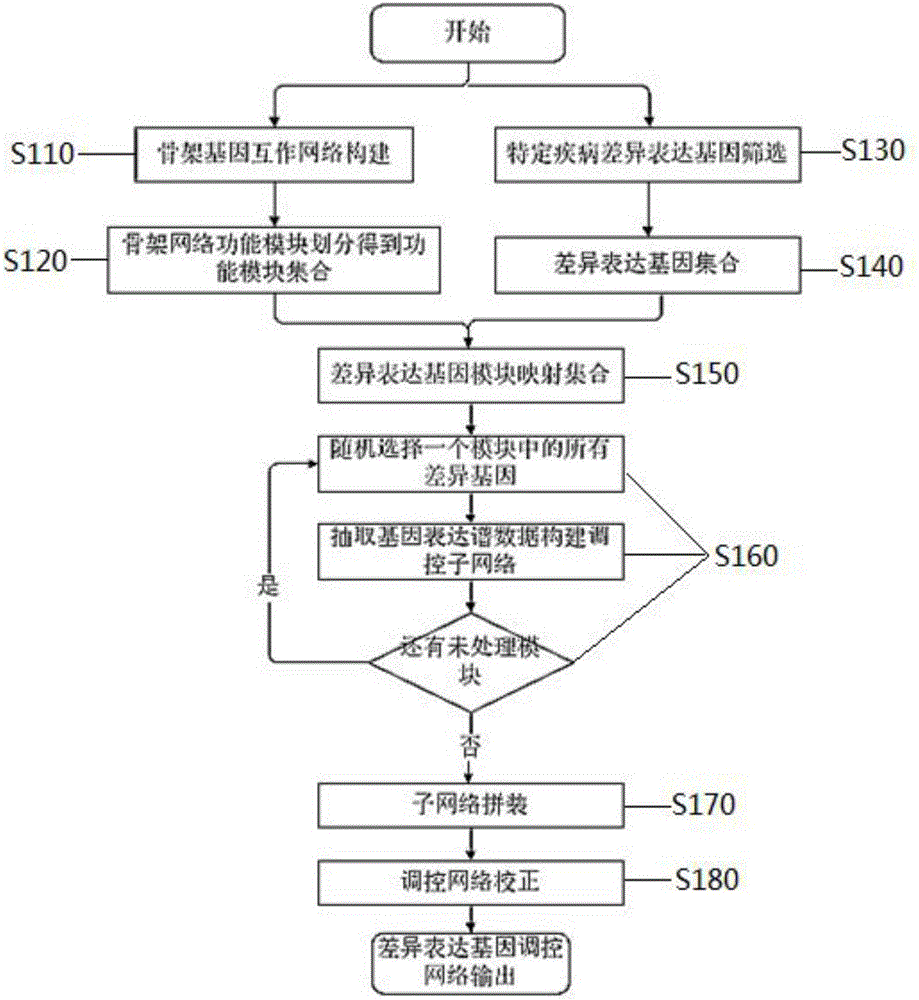
Construction method for specific cancer differential expression gene regulation and control network
InactiveCN105740651AHigh precisionImprove calculation accuracyHybridisationSpecial data processing applicationsDifferentially expressed genesBioinformatics
The invention discloses a construction method for a specific cancer differential expression gene regulation and control network.The method includes the following steps of firstly, constructing a framework gene interaction network according to function similarity weight numbers between genes; secondly, conducting module division on the framework gene interaction network through a segmenting method; thirdly, screening out differential expression genes through complete genome methylation data; fourthly, classifying the screened-out differential expression genes according to functions; fifthly, using all the differential expression genes mapped to each same function module as a function classification; sixthly, constructing the regulation and control network of all the genes in each function classification function; seventhly, conducting sub-network splicing under guidance of a framework network.The calculation complexity is greatly reduced, and high precision is achieved.
Owner:JILIN UNIV
Gene regulation and control network constructing method based on Bayesian network
InactiveCN101763528APrecise representation structureIndicates the structureGenetic modelsNODALLocal optimum
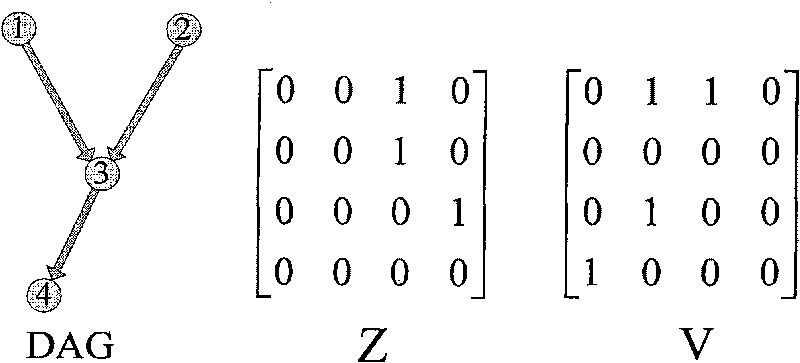
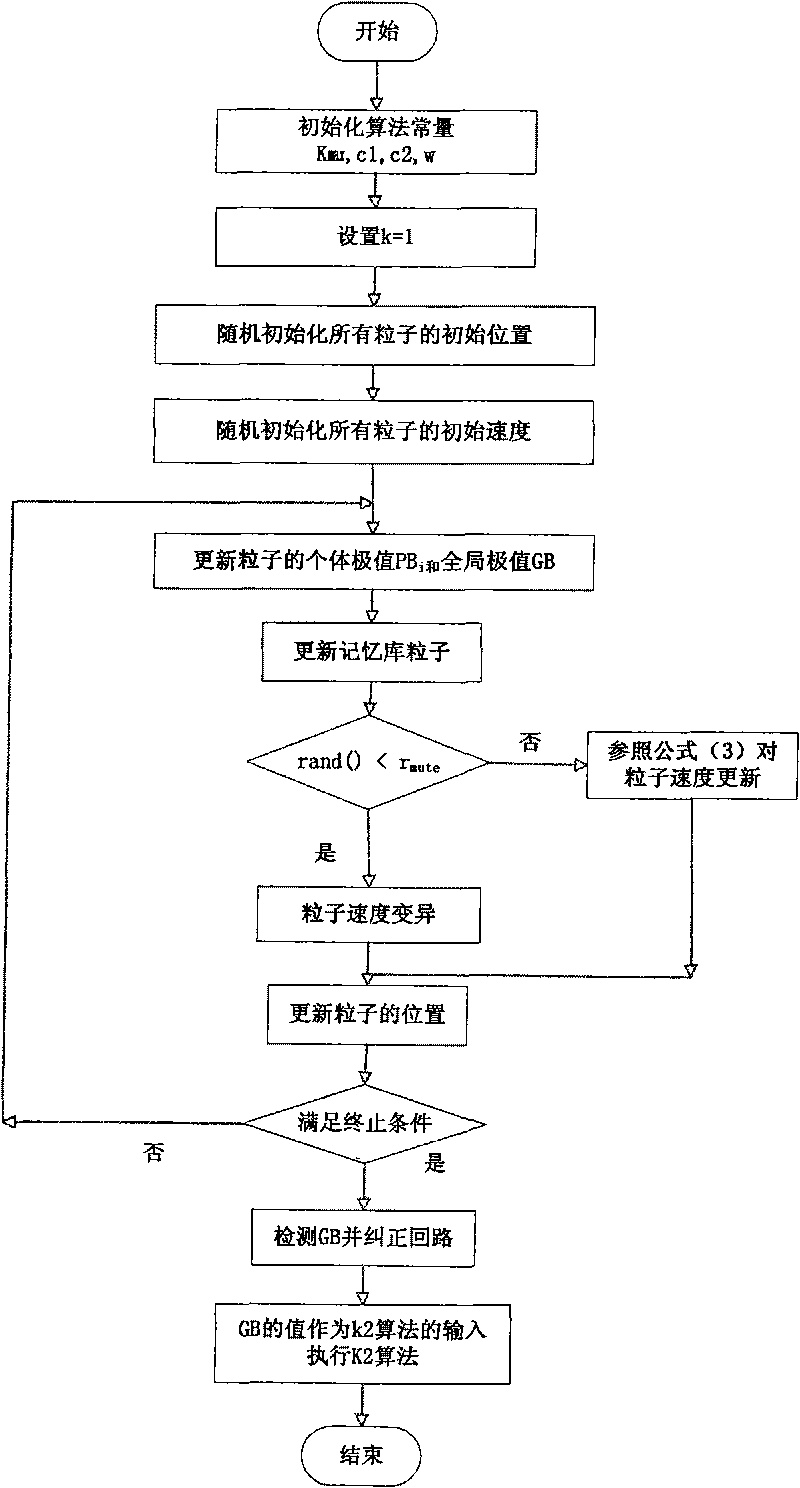
The invention relates to a gene regulation and control network constructing method based on a Bayesian network. Relationship of all gene expressions in a species or a tissue is wholly analyzed and studied in simulation by constructing a gene regulation and control network model. The construction of the gene regulation and control network model comprises the following steps: A. identifying the best node order by a binary particle swarm optimization algorithm with memories, wherein after the particle speed is updated by the binary particle swarm optimization algorithm, the speed of part particles is varied and a searching space is searched, and skipping from local is taken as optimal; and B. inputting the obtained best node order as a K2 algorithm, then executing K2 algorithm and studying the structure of the Bayesian network. The method has the advantages of fast convergence, simple calculation, easy realization and the like, provides clues for studying regulation and control of gene transcription level, shows the structure of the gene regulation and control network more accurately and has important theoretical significance and practical value in many fields, such as biology, medicine science, pharmacy and the like.
Owner:SHENZHEN UNIV
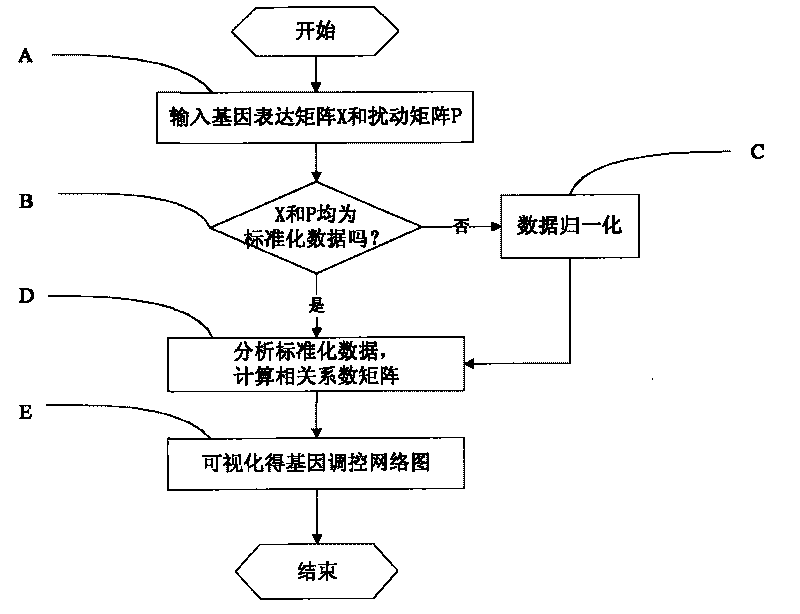
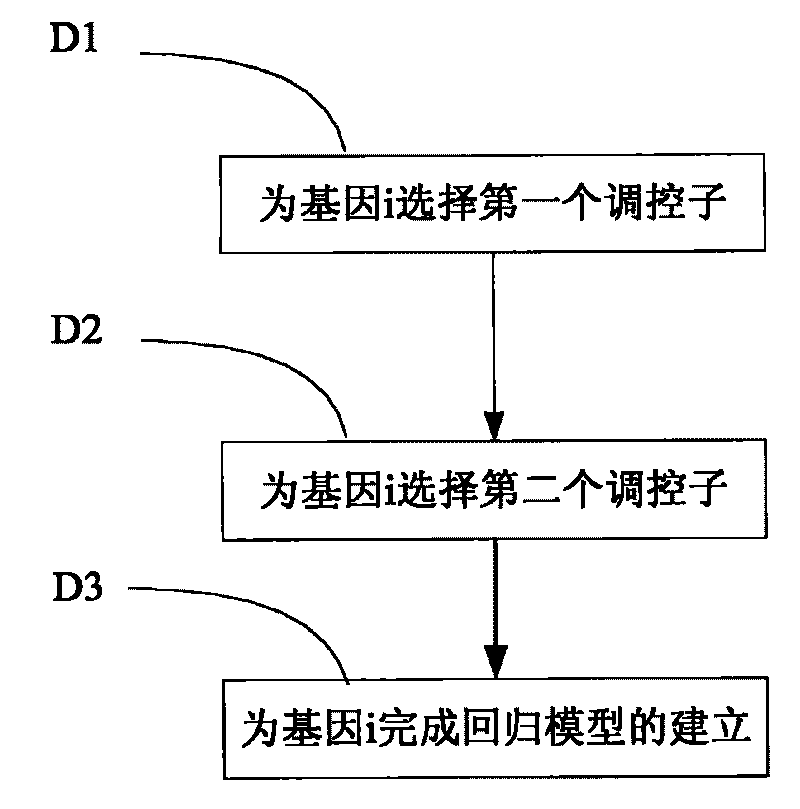
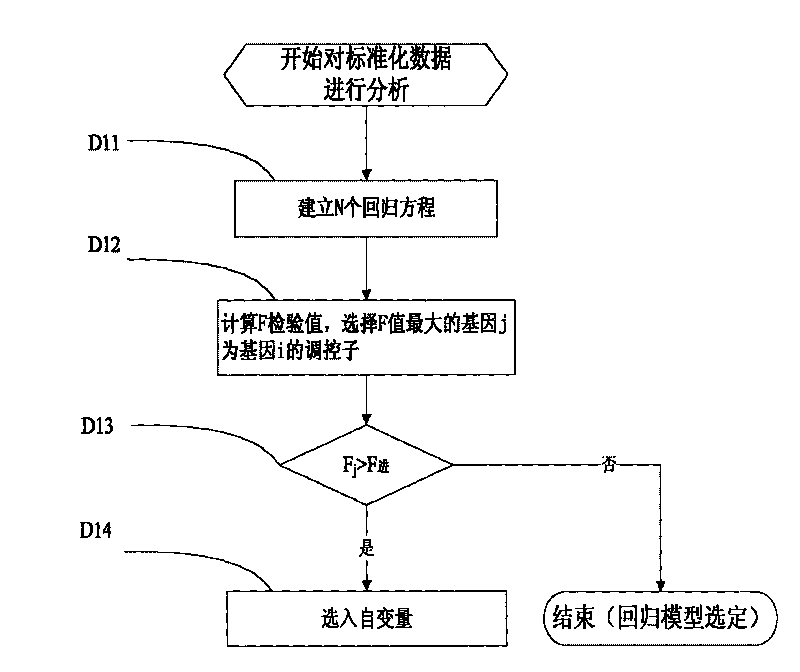
Inference method of stepwise regression gene regulatory network
InactiveCN101719195AOvercoming Experimental Data ProblemsExperimental data problem solvingSpecial data processing applicationsSmall sampleAssay standardization
The invention discloses an inference method of a stepwise regression gene regulatory network. The method comprises the following steps of: A, reading a gene expression data matrix and a gene perturbation data matrix; B, confirming whether the gene expression data matrix and the gene perturbation data matrix are standardized data or not; C, respectively carrying out data normalization on the gene expression data matrix and the gene perturbation data matrix to form the standardized data; D, analyzing the standardized data and calculating all inter-gene correlation coefficient matrixes; and E, visualizing the inter-gene correlation coefficient matrixes into a network to obtain a gene regulatory network chart. The method can select optimal regression subsets to solve the problem of high-dimension small-sample experimental data, gradually select the most influential regulator for a target gene, accord with the true condition of the gene regulatory network, and be superior to similar methods in calculation precision and calculation efficiency along with the enlargement of the gene regulatory network scale and the network sparsity.
Owner:SHANGHAI UNIV
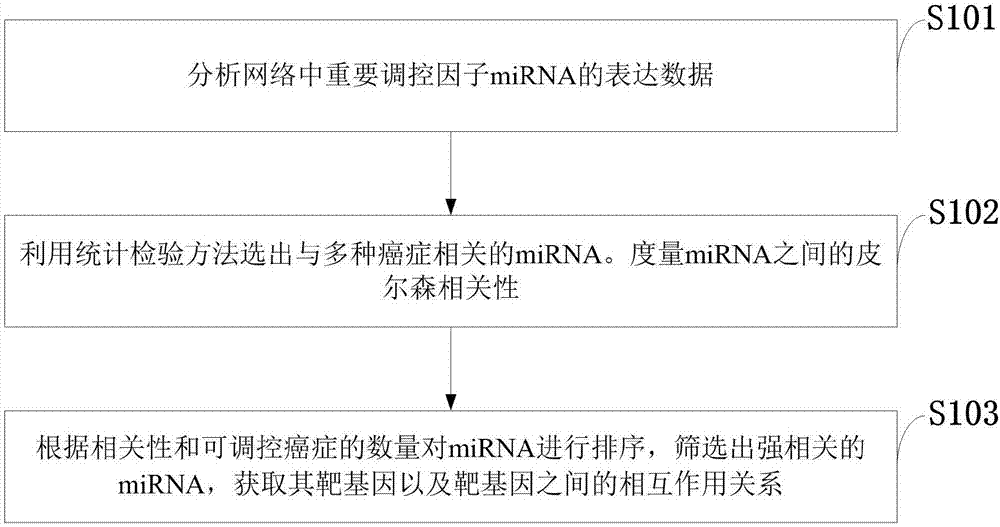
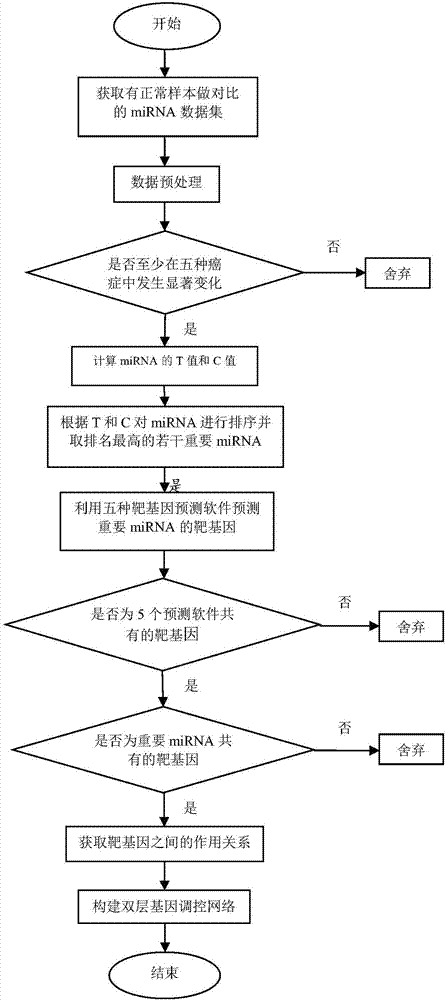
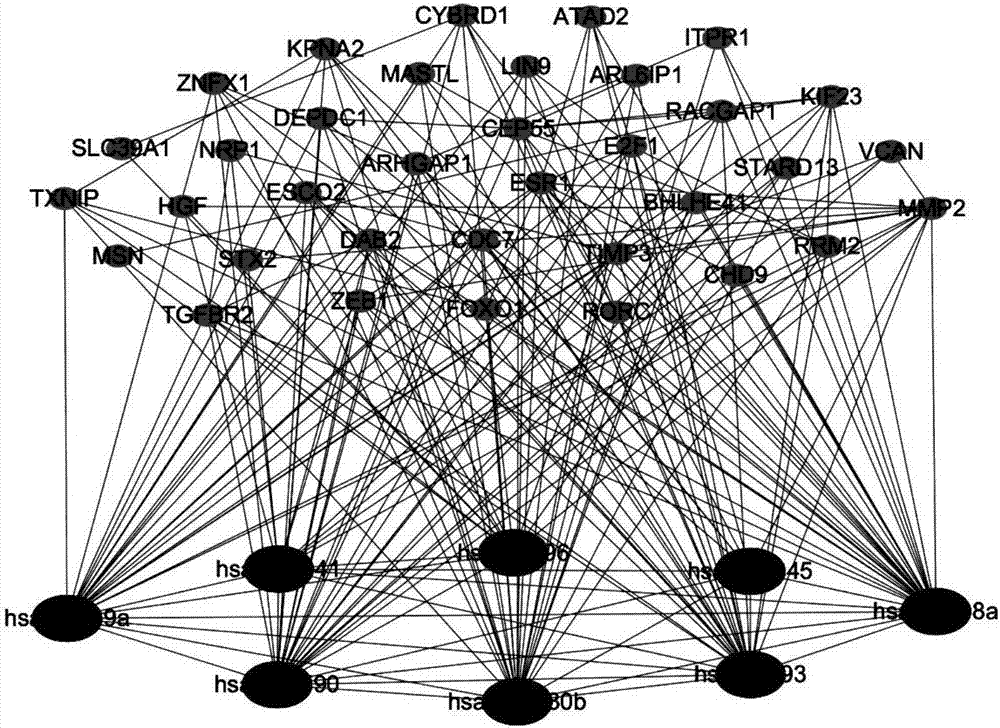
Method for constructing double-layer gene regulation and control network
ActiveCN107358062AUnderstanding PathogenesisUnderstand the processProteomicsGenomicsKey factorsTargeted therapy
The invention belongs to the technical field of data processing and discloses a method for constructing a double-layer gene regulation and control network. The method comprises following steps: analyzing expression data of an important regulation and control factor miRNA in a network; selecting miRNA relevant to multiple types of cancer by means of the statistics examination method; measuring Pearson correlation among the miRNA; ordering the miRNA according to the correlation and the number of controllable cancer, screening out strongly correlated miRNA and obtaining target genes thereof and interacting relations among the target genes. The present invention discloses the mechanism of gene and miRNA involved in the regulation of biological processes such as cancer and helps to understand the relationship between genes and cancer and other complex diseases and provides reference for the development of biopharmaceuticals and targeted therapy for pan-cancer. Identifying cancer-associated miRNAs and genes can be used to elucidate the mechanisms of cooperation that are involved in the regulation of key factors that influence a variety of cancer processes, cancer risk prediction, and the development of bio-targeted drugs.
Owner:XIDIAN UNIV
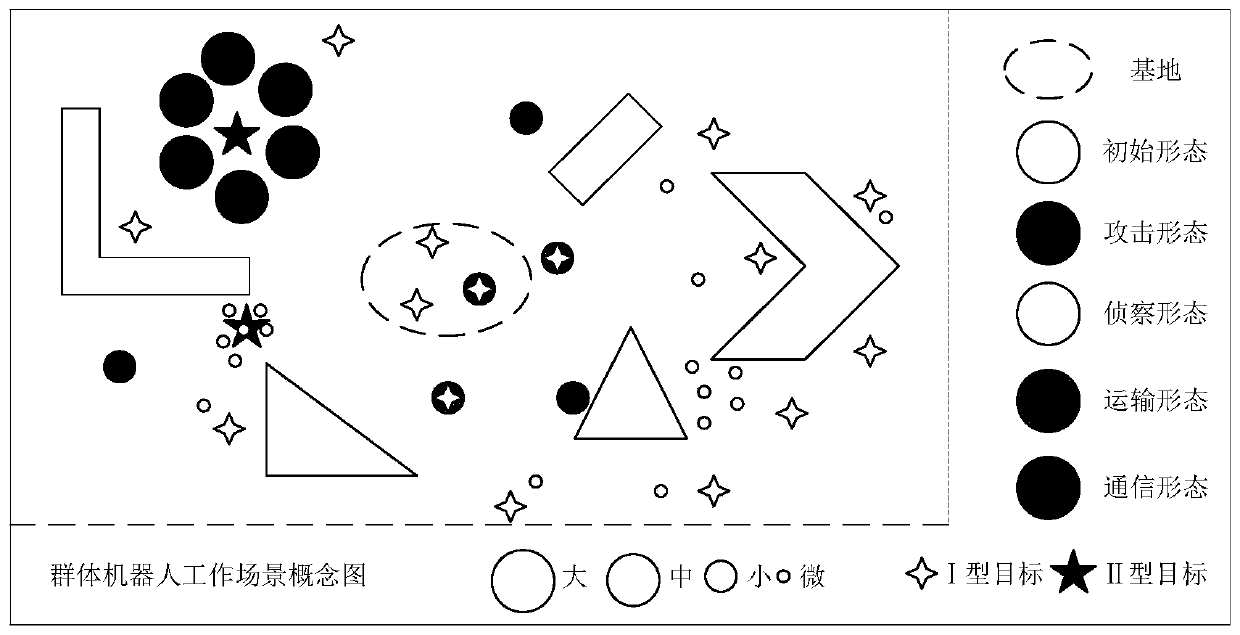
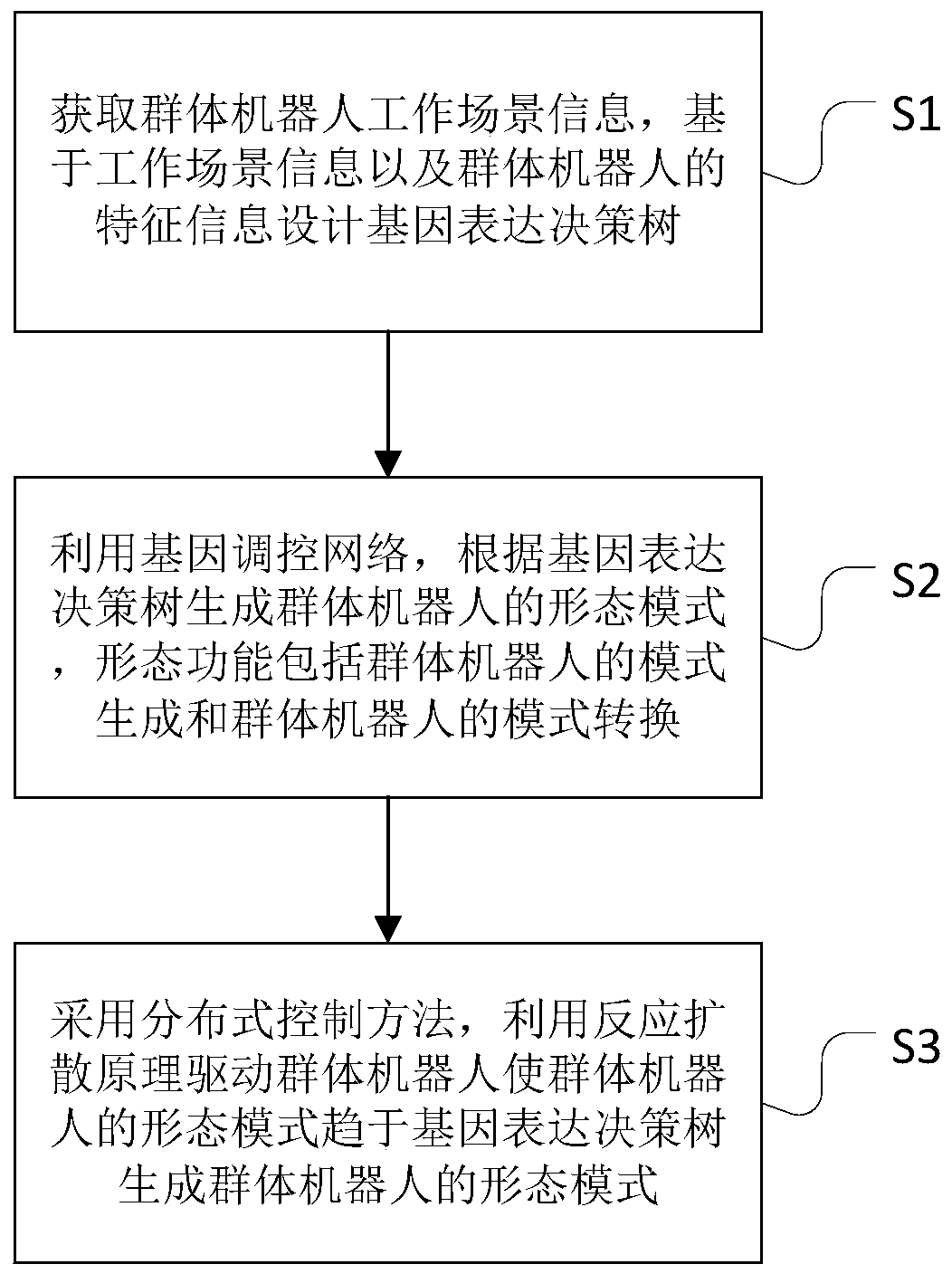
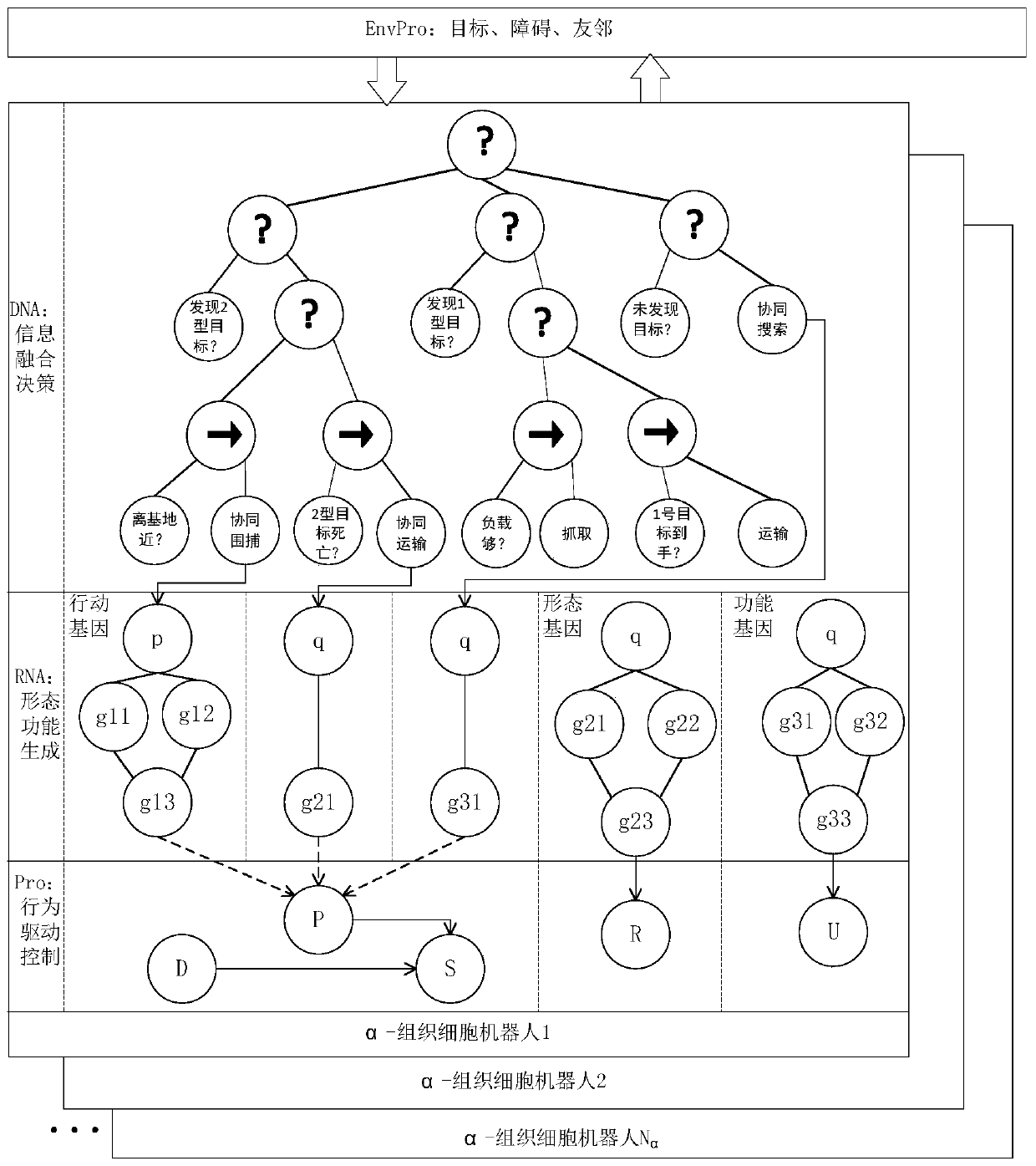
Swarm robot mode generation and conversion method of multistage variable gene regulation and control network
ActiveCN111476337AOptimal Morphological DecisionValid conversionArtificial lifeExpression geneControl theory
The invention provides a swarm robot mode generation and conversion method for a multistage variable gene regulation and control network, and the method comprises the following steps: obtaining the work scene information of a swarm robot, and designing a gene expression decision tree based on the work scene information and the feature information of the swarm robot; generating a morphological modeof the swarm robot according to the gene expression decision tree by utilizing a gene regulation and control network, wherein morphological functions comprise mode generation of the swarm robot and mode conversion of the swarm robot; adopting a distributed control method, and employing a reaction diffusion principle for driving the swarm robot to enable the morphological mode of the swarm robot to tend to a gene expression decision tree to generate the morphological mode of the swarm robot. According to the method, an optimal form decision can be made in a changing environment, and form generation and conversion can be effectively completed.
Owner:NAT UNIV OF DEFENSE TECH
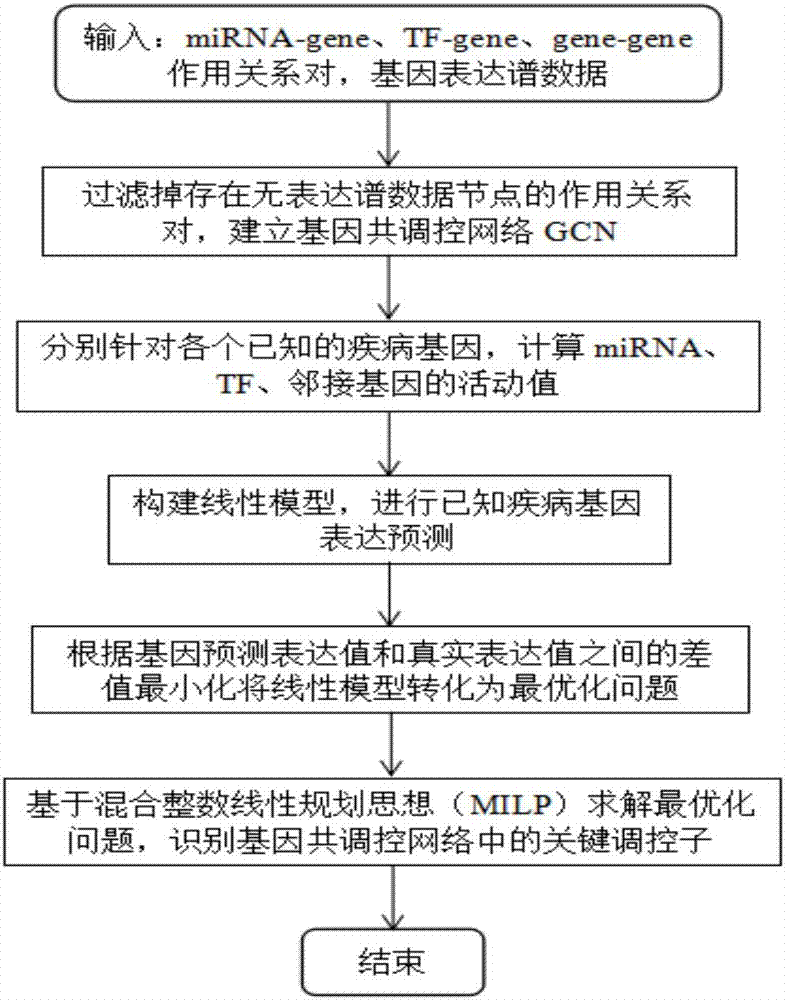
Method for recognizing key regulator in gene co-regulatory network based on linear model
ActiveCN106874704AHigh precisionAccurate identificationBiostatisticsSystems biologyLinear modelGene expression profiling
The invention discloses a method for recognizing a key regulator in a gene co-regulatory network based on a linear model. The gene expression profile data and the gene regulation relation data are utilized to forecast the expression of the known disease gene in the manner of establishing the linear model, so as to complete the recognition for the key regulator in the gene co-regulatory network. The method provided by the invention is simply realized; the key regulator in the gene co-regulatory network can be accurately recognized only according to the gene expression profile data and the gene regulation relation; tests prove that the recognized regulator has an important biological significance and has important theoretical significance and practical value in researching the disease mechanism.
Owner:HUNAN UNIV
MiRNA molecules isolated from human embryonic stem cell
ActiveUS7674617B2Good curative effectSugar derivativesMicrobiological testing/measurementDevelopmental stageMammal
The present invention relates to novel miRNA molecules, more particularly to novel miRNA molecules isolated from human embryonic stem cells. The miRNA molecules provided by the present invention can be usefully used as a molecular marker for early developmental stages of undifferentiated human embryonic stem cells. Also, the miRNA molecules of the present invention may play an important role in the regulation of mammalian embryonic stem cells. Therefore, the miRNA molecules can be usefully used for analyzing regulatory networks of human embryonic stem cells.
Owner:COLLEGE OF MEDICINE POCHON CHA UNIV IND
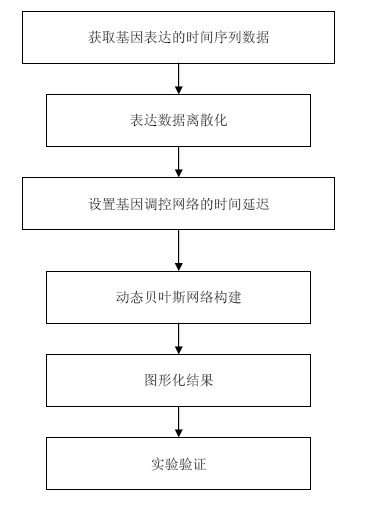
Gene regulatory network constructing method based on dynamic Bayesian network
InactiveCN102013039AReduce complexityPromote reconstructionGenetic modelsStructure learningDiscretization
The invention relates to a gene regulatory network constructing method based on a dynamic Bayesian network. The method comprises the following steps: 1) the time series data of gene expression can be obtained; 2) the discretization method is adopted to discretize the time series data into a plurality of expression levels; 3) the time delay of the gene regulatory network is set; and 4) the structure learning algorithm of the dynamic Bayesian network is utilized and the maximum likelihood method is adopted to deduce the gene regulatory network. By adopting the gene regulatory network constructing method, the complexity can be reduced effectively, the reconstruction performance can be improved; and the method has high practicability.
Owner:HANGZHOU NORMAL UNIVERSITY
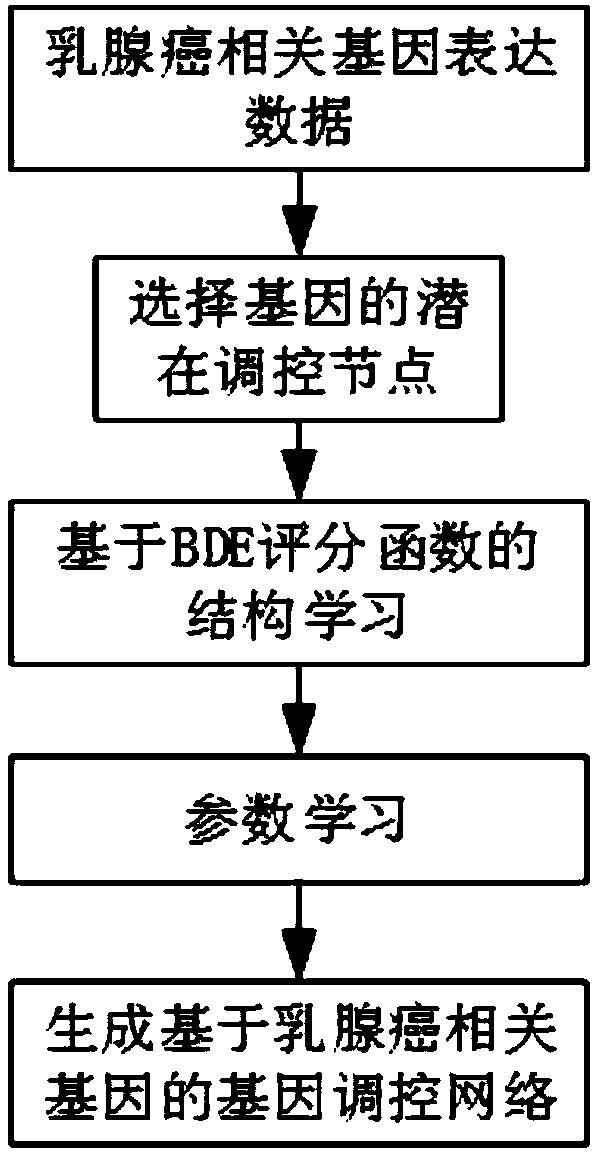
Regulatory network construction and analysis method based on breast cancer diseases
The invention provides a gene regulatory network construction and analysis method based on breast cancer diseases. The flow comprises the steps: preliminary screening of breast cancer-related genes; construction of the gene regulatory network; performing node centrality analysis according to the result of the gene regulatory network, including node degree centrality, approximation centrality, intermediary centrality and eigenvector centrality; and taking the first N data of network node degree centrality, approximation centrality, intermediary centrality and eigenvector centrality as the selected related genes. The breast cancer-related genes are effectively selected out from human genes so as to establish the gene regulatory network related to the breast cancer diseases, important genes are obtained through node centrality analysis, the research on the breast cancer diseases from the perspective of genetics can be promoted and a foundation can be laid for finding an effective way to intervene the breast cancer diseases.
Owner:NORTHEASTERN UNIV LIAONING
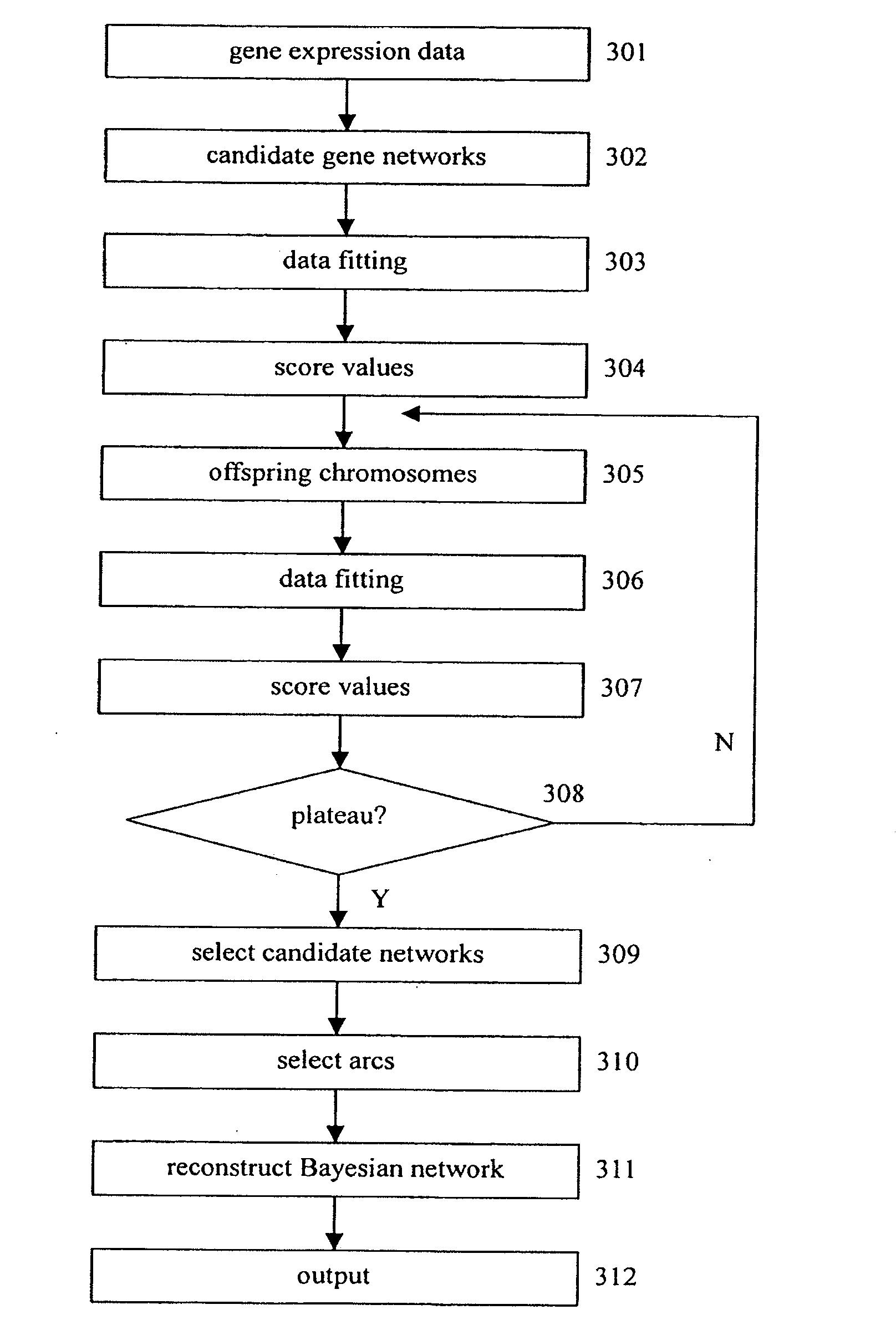
Reconstruction of gene networks from time-series microarray data
InactiveUS20050256652A1Microbiological testing/measurementDigital computer detailsCandidate Gene Association StudySimplex algorithm
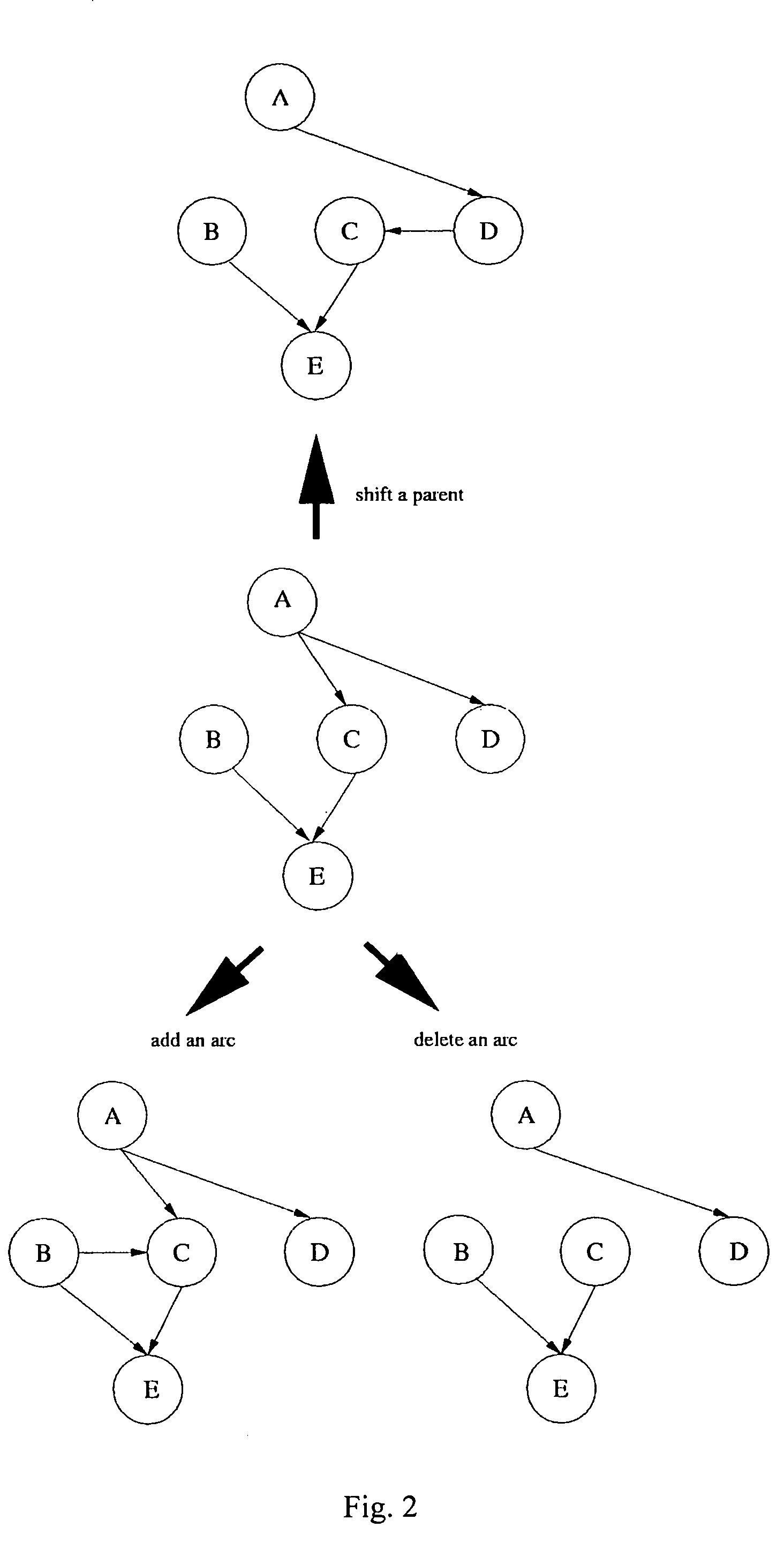
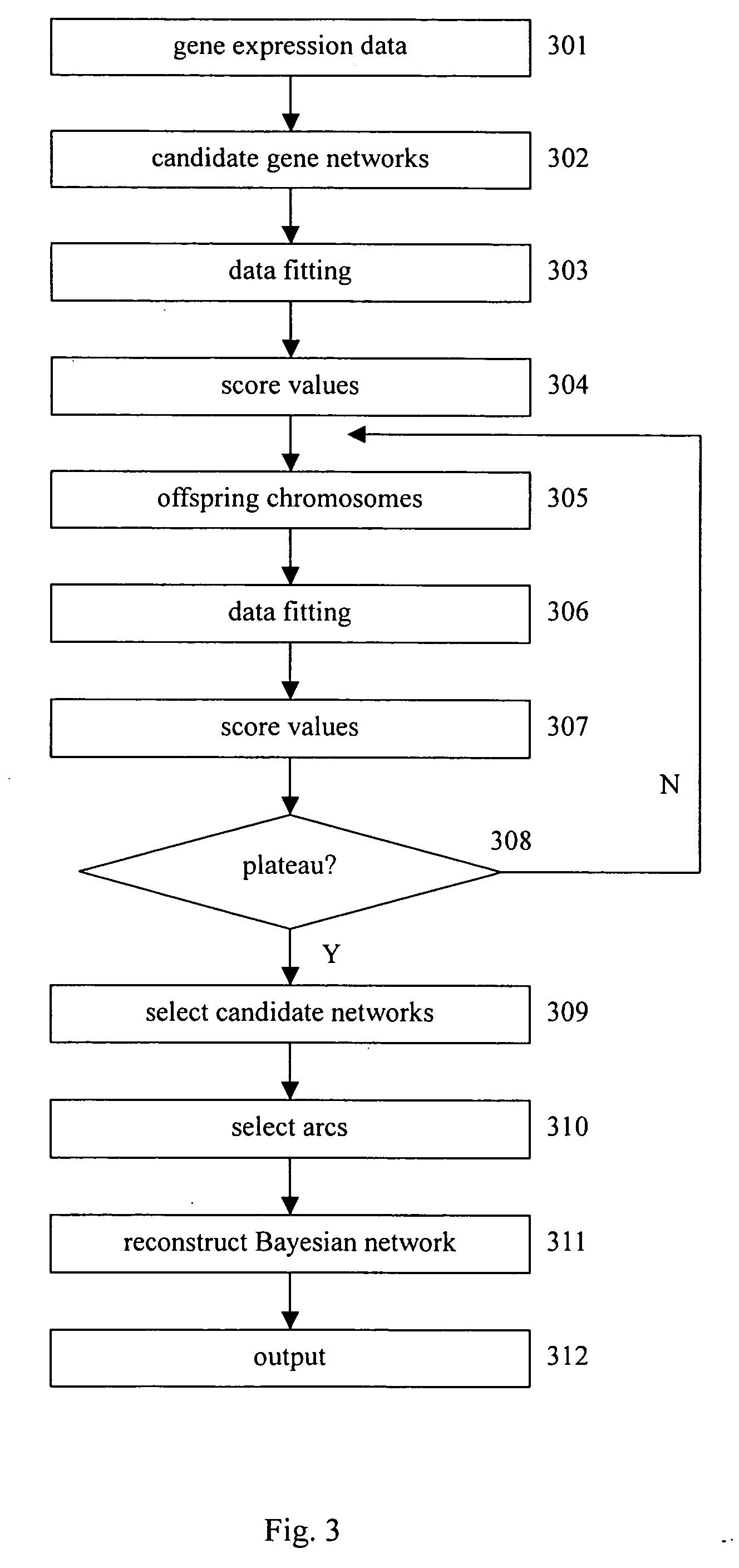
Gene regulation network is reconstructed using time series microarray data under the method of the Bayesian network. Particular power-law function is used to calculate the joint probabilities among genes across time points. This invention discloses the use of the downhill simplex algorithm to find global maxima of interrelational likelihood. Arcs with higher frequencies are selected to establish the gene regulation network. Prior knowledge may be included into candidate gene networks to accelerate search for best networks.
Owner:ACAD SINIC
PhoP deleted riemerella anatipestifer CH1 attenuated strain and construction method thereof
InactiveCN104560855ALittle knowledgeAntibacterial agentsBacterial antigen ingredientsMicrobiologyRiemerella anatipestifer
The invention discloses a riemerella anatipestifer CH1 global regulator phoP attenuated strain and a construction method thereof, and belongs to the technical field of biological engineering. The attenuated strain is constructed by knocking out a phoP gene by means of a homologous recombination method and utilizing a bacterial conjugation method. The prepared attenuated strain has a preservation number of CCTCC NO:M2014557, and is preserved on November 7, 2014. The attenuated strain has low toxicity, and a foundation is established for construction of subsequent vaccine vectors and construction of a riemerella anatipestifer gene regulatory network. The invention further provides application of the attenuated strain in vaccines.
Owner:SICHUAN AGRI UNIV
Conjoined grafting method for plant meristem
The invention relates to a conjoined grafting method for plant meristem. The conjoined grafting method for the plant meristem is characterized in that the conjoined grafting method through meristem between different phylum, klasse, order, family, genus and species of plantae is provided, and the conjoined grafting method comprises an apcial meristem, a lateral meristem and an intercalary meristem. The meristem possesses massive reversible gene regulatory networks and spatial and temporal expression opportunities due to the fact that the meristem has high cell division ability and is a key tissue of organogenesis and morphogenesis and a starting point of organ and tissue differentiation. Therefore, conducting moderate manual injury intervention and conjoined grafting on the meristem can greatly increase opportunity of successful grafting, and enlarges boundary capable of being conducted grafting between different phylum, klasse, order, family, genus and species of the plantae. Due to the fact that scion and rootstock whole plant are not separated, bud mutation and fructification are promoted to obtain grafting progeny seeds, exchanging and changing of part genetic materials of grafting progeny can be achieved, and consequently, low coat and safe material exchanging of extra distant species of the plantae can be achieved, variation is induced, and then a new germplasm of a scion plant is created.
Owner:YANGTZE UNIVERSITY
Three-component crispr/cas complex system and uses thereof
ActiveUS20180094257A1Lack nuclease activityPolypeptide with localisation/targeting motifFusion with RNA-binding domainGenomeDNA
The invention described herein provides compositions and reagents for assembling a tripartite complex at a specific location of a target DNA. The invention also provides methods for using the complex to, for example, label a specific genomic locus, to regulate the expression of a target gene, or to create a gene regulatory network.
Owner:JACKSON LAB THE
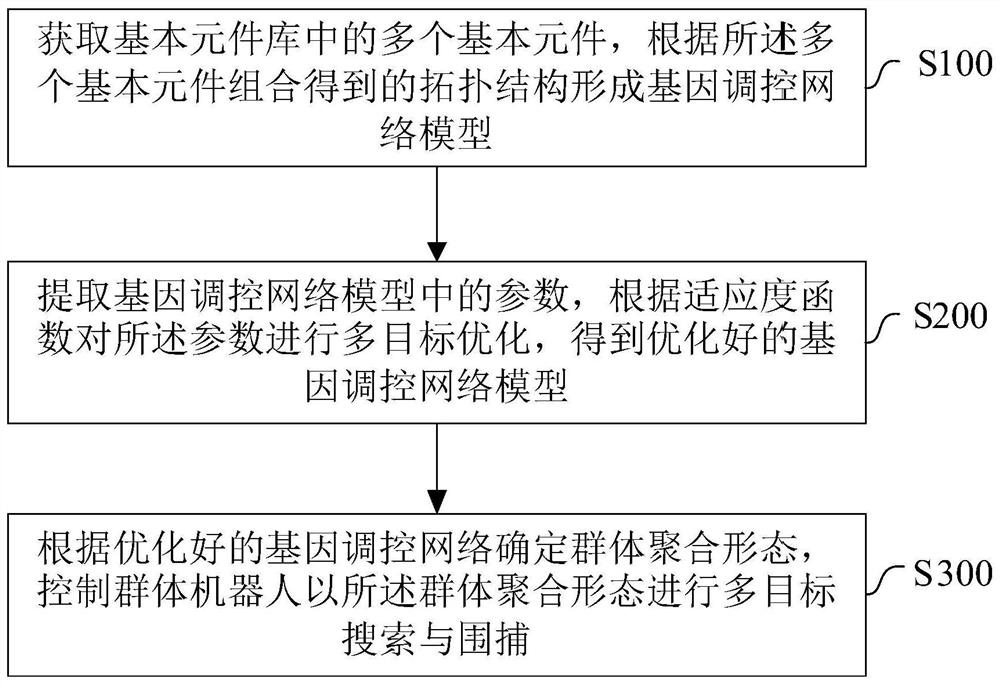
Multi-target searching and capturing control method and system for swarm robots
The invention relates to the technical field of swarm robot control, in particular to a multi-target searching and capturing control method and system for swarm robots, and the method comprises the steps: obtaining a plurality of basic elements in a basic element library, and forming a gene regulation and control network model according to a topological structure obtained through the combination of the plurality of basic elements; extracting parameters in the gene regulation and control network model, and performing multi-objective optimization on the parameters according to a fitness function to obtain an optimized gene regulation and control network model; determining the group aggregation form according to the optimized gene regulation and control network, andcontrolling the group robot to perform multi-target searching and capturing in the group aggregation form. Multiple targets can be captured in a complex dynamic environment, and the method has good adaptability.
Owner:SHANTOU UNIV
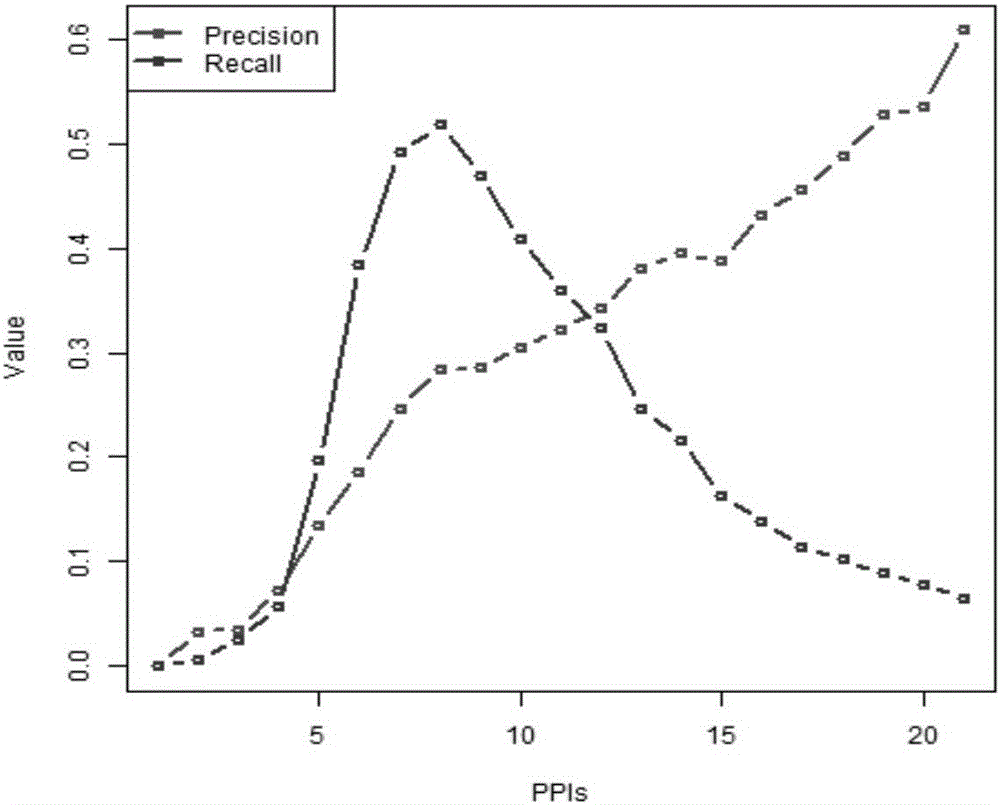
Gene regulation and control network reconstruction method based on gene expression data
ActiveCN108197432AImprove accuracyEasy to moveHybridisationSpecial data processing applicationsDifferential evolutionGene regulatory network
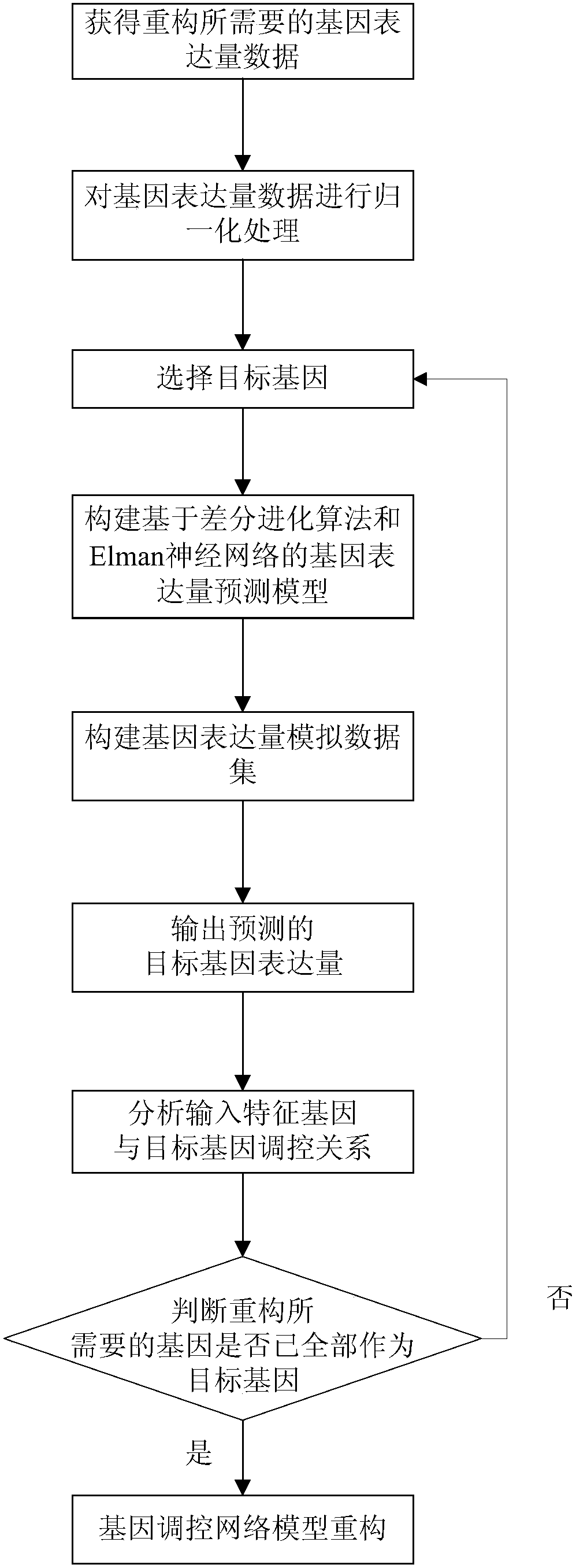
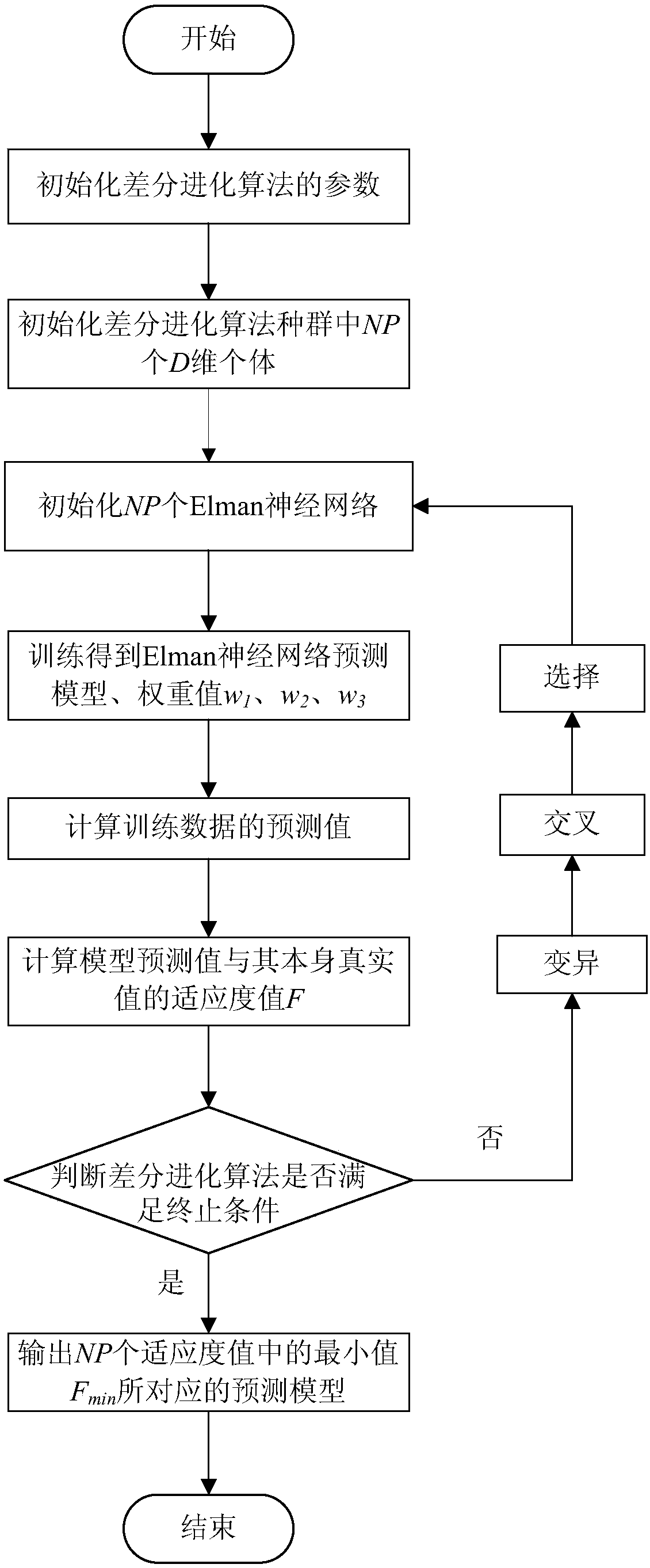
The invention provides a gene regulation and control network reconstruction method based on gene expression data, and relates to the technical field of gene regulation and control network reconstruction in bioinformatics. The method comprises the following steps that: obtaining gene expression quantity data required for reconstruction; carrying out normalization processing on the data; carrying out predictive modeling on a target gene expression quantity; predicting the target gene expression quantity; analyzing a regulation and control relationship between an input feature gene and a target gene; and reconstructing the gene regulation and control network. By use of the gene regulation and control network reconstruction method based on the gene expression data, high-accuracy gene regulation and control network modeling is realized according to gene expression data, and an Elman neural network optimized by a differential evolution algorithm predicts the gene expression quantity so as tohave the advantages of high operation speed and high accuracy. In addition, simulation data can be used for solving the problem of an insufficient data quantity is solved, the gene regulation and control network which is finally established exhibits good accuracy, has a wide applicable range, can be suitable for different pieces of gene expression data and exhibits good transportability.
Owner:NORTHEAST DIANLI UNIVERSITY
Gene regulation and control network re-building method based on cross-platform gene expression data
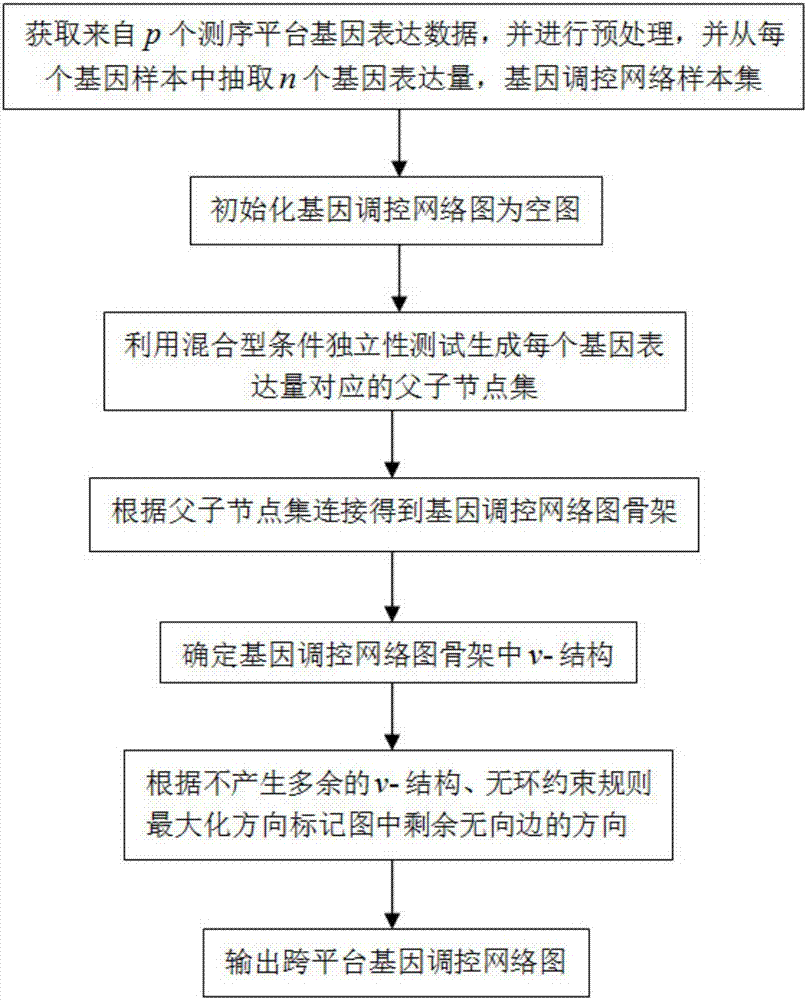
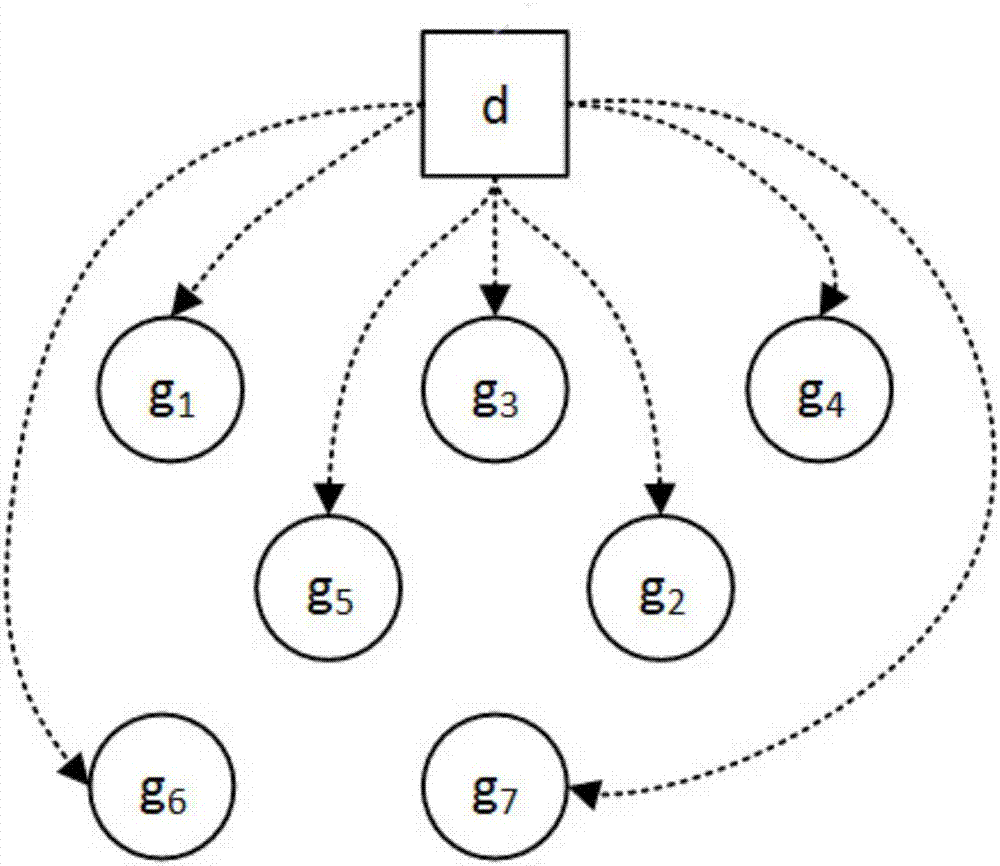
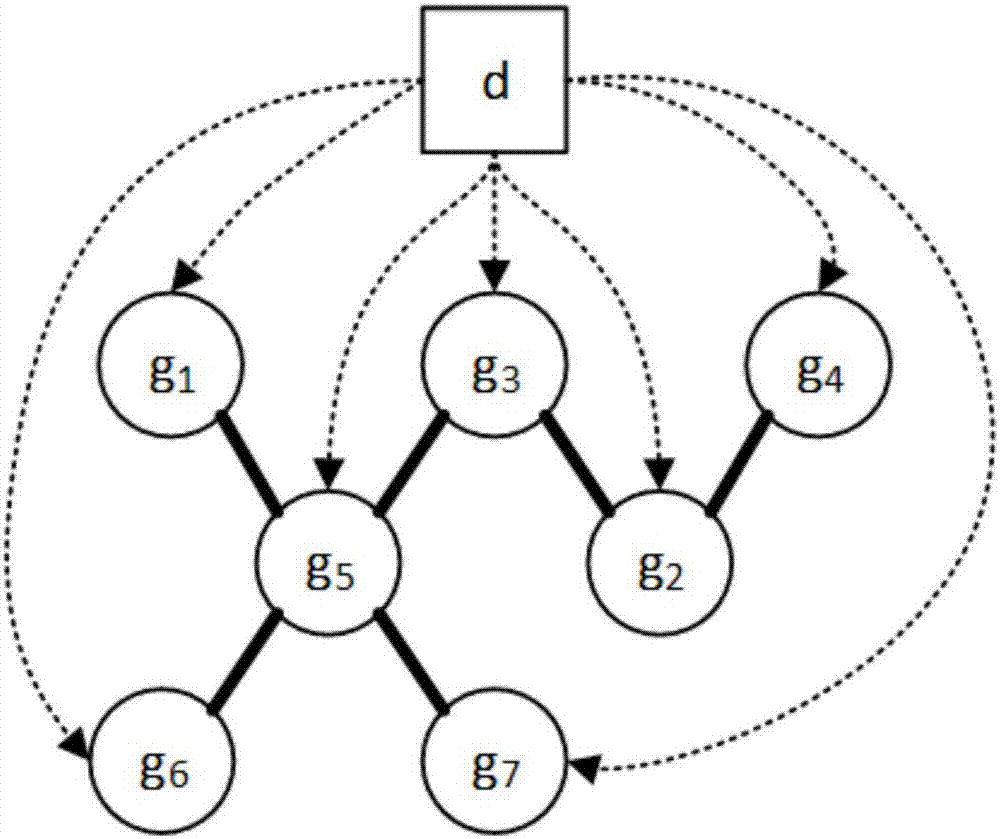
ActiveCN107016260ASolve poor comparabilitySolve high-dimensional problemsSpecial data processing applicationsNODALHybrid type
The invention relates to a gene regulation and control network re-building method based on cross-platform gene expression data. Gene expression data from p sequencing platforms is obtained and preprocessed according to the characteristics of the cross-platform gene expression data, n gene expression quantities are extracted from each gene sample, a father and son node sets of each gene expression quantity is obtained based on the mixed type condition independence testing of partial correlation coefficients, the father and son node sets are applied to the three processes of a learning network framework of cross-platform cause-effect network structure learning, v-structure determination and maximized direction marking, and a cross-platform gene regulation and control network is rebuilt. The problems of a cross-platform gene regulation and control network are solved through a cause-effect graph model, high-dimensional gene regulation and control network rebuilding can be directly and effectively carried out through the cross-platform gene expression data, data excess smoothness and other problems caused in the data preprocessing process are avoided, and the correctness rate and the recall rate of the rebuilding result of the cross-platform gene regulation and control network are increased.
Owner:GUANGDONG UNIV OF TECH
Reconstruction of gene networks from time-series microarray data
InactiveUS20090063376A1Genetic modelsDigital computer detailsCandidate Gene Association StudySimplex algorithm
Gene regulation network is reconstructed using time series microarray data under the method of the Bayesian network. Particular power-law function is used to calculate the joint probabilities among genes across time points. This invention discloses the use of the downhill simplex algorithm to find global maxima of interrelational likelihood. Arcs with higher frequencies are selected to establish the gene regulation network. Prior knowledge may be included into candidate gene networks to accelerate search for best networks.
Owner:ACAD SINIC
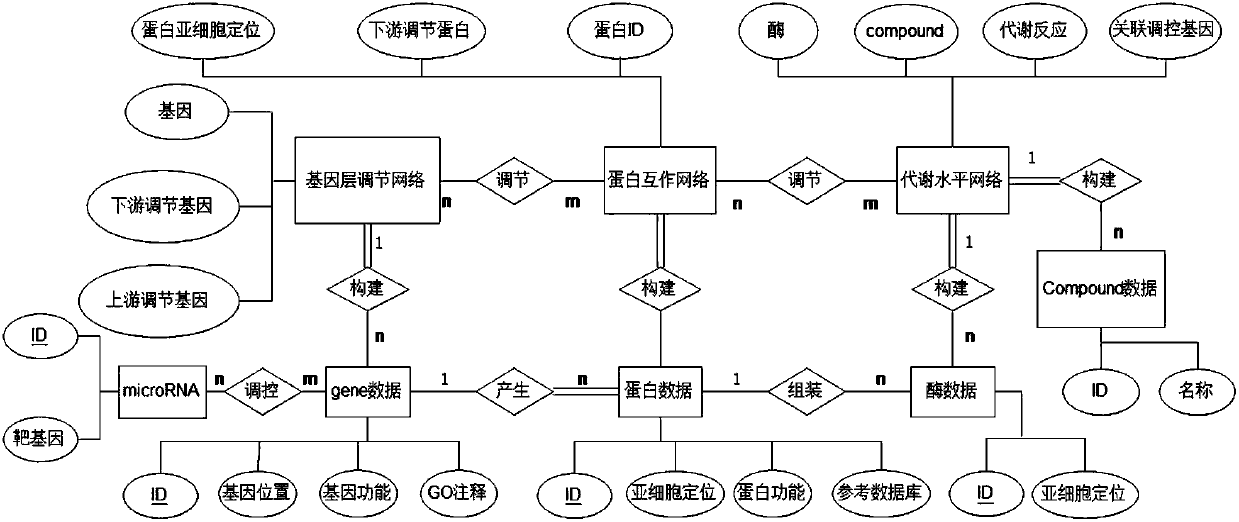
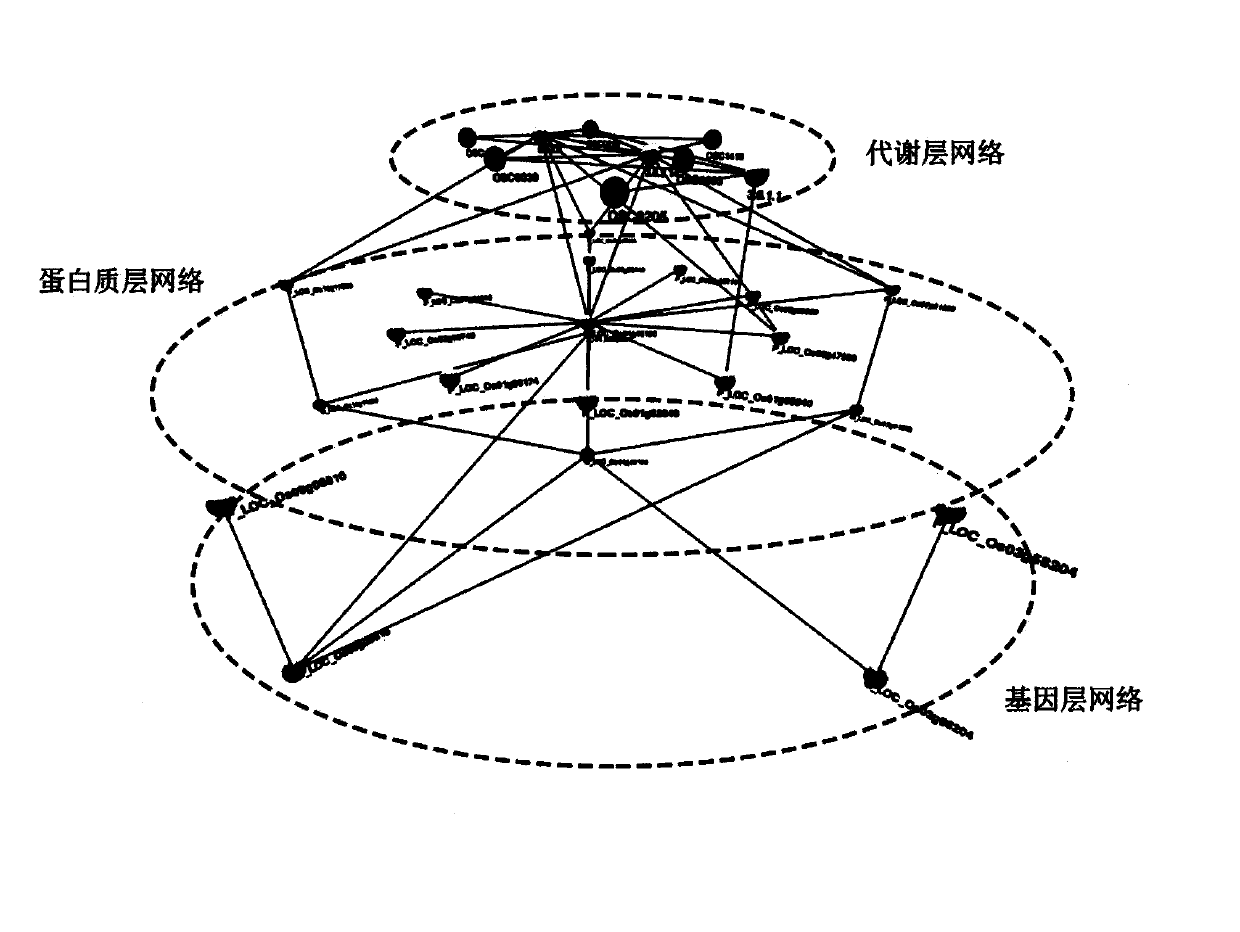
Plant whole-genome multilevel biological network reconstruction method based on multi-omics data integration
InactiveCN107862176ARealize 3D visualizationSpecial data processing applicationsReconstruction methodMetabolic network
The invention discloses a plant whole-genome multilevel biological network reconstruction method based on multi-omics data integration. The method carries out integration on a rice gene regulation andcontrol network, a protein interaction network and a metabolic network to finish the construction of a rice multilevel regulation and control network; and meanwhile, through the integration of expression profile data in different tissue growth processes of the rice, according to the constructed rice multilevel regulation and control network, a specific tissue multilevel gene regulation and control network is constructed so as to construct a first high-quality rice genome scale multilevel biological network of which the current data level is most integral. A new thought is provided for the construction of the plant multilevel biological network, and a constructed rice multilevel gene regulation and control network database realizes the 3D (Three-Dimensional) visualization of the multilevelregulation and control network.
Owner:ZHEJIANG UNIV
Inferring gene regulatory networks from time-ordered gene expression data using differential equations
Embodiments of methods are provided that can be used to estimate network relationships between genes of an organism using time course expression data and a set of linear differential equations. Aikaike's Information Criterion and mask tools can be used to reduce the number of elements in a matrix by determining which elements are zero or not significantly changed under the conditions of the study. Maximum likelihood estimation and new statistical methods are used to evaluate the significance of a proposed network relationship.
Owner:GNI USA +1
Method for constructing mRNA 5'-termnal information library
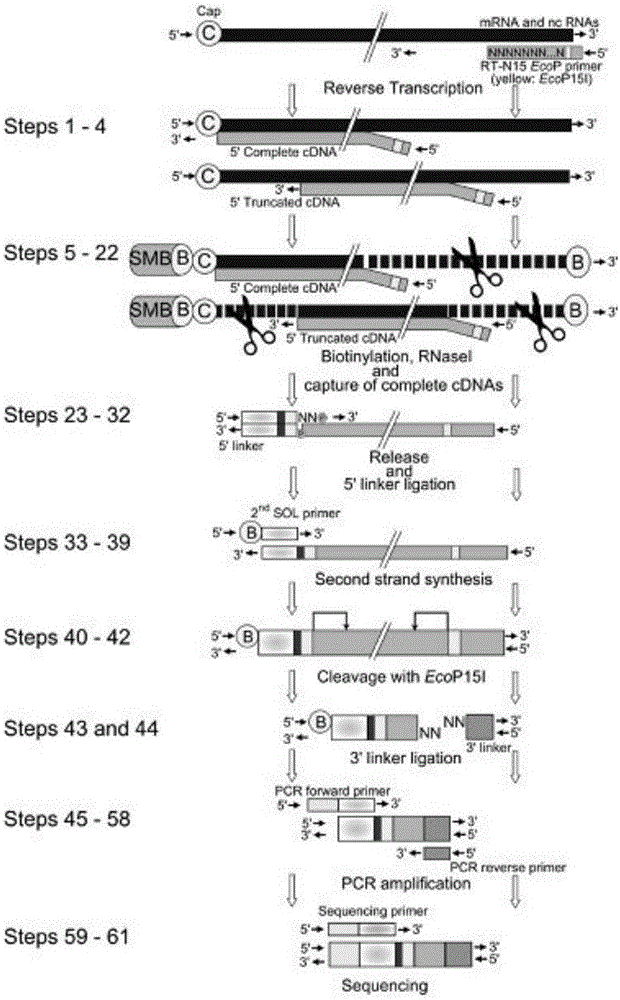
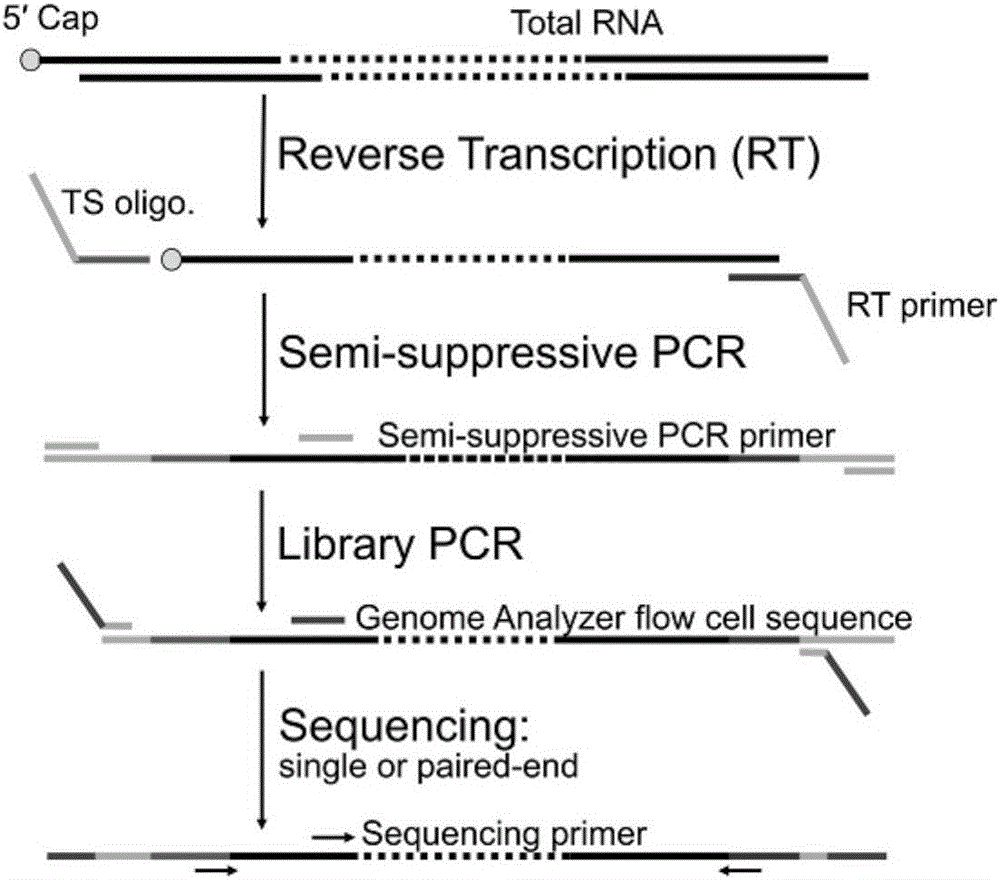
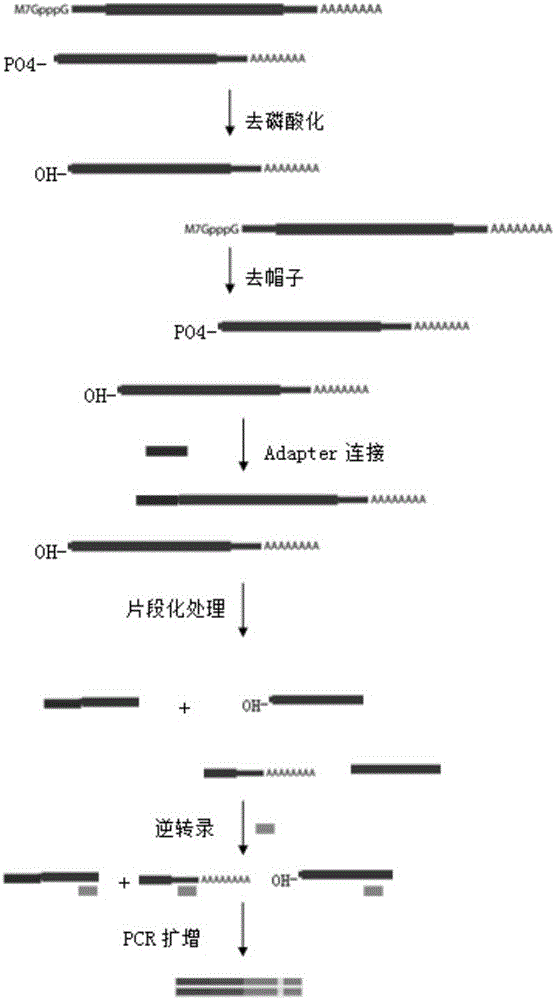
ActiveCN105734052APromote enrichmentShorten the lengthMicrobiological testing/measurementLibrary creationGenomicsStart site
The invention belongs to the technical field of genomics, and particularly relate to a method for simplifying CAGE-seq library establishing.The method comprises the steps that mRNA and lncRNA containing cap structures are enriched, a 5'-termnal connector is specifically added, after fragmentation processing, reverse transcription is carried out, and 5'-terminal information is further enriched.According to the whole method, an existing mature RNA-seq kit of the gnomegen company is mixed.The method is easier to operate and high in success rate, the 5'-terminal information can be specifically enriched, the method is more helpful for verifying the transcriptional start site of mRNA in a whole cell, and thus acquiring accurate promoter information and 5'UTR information.The method is a powerful mode for studying gene expressions and is suitable for gene regulatory network studies.
Owner:武汉生命之美科技有限公司
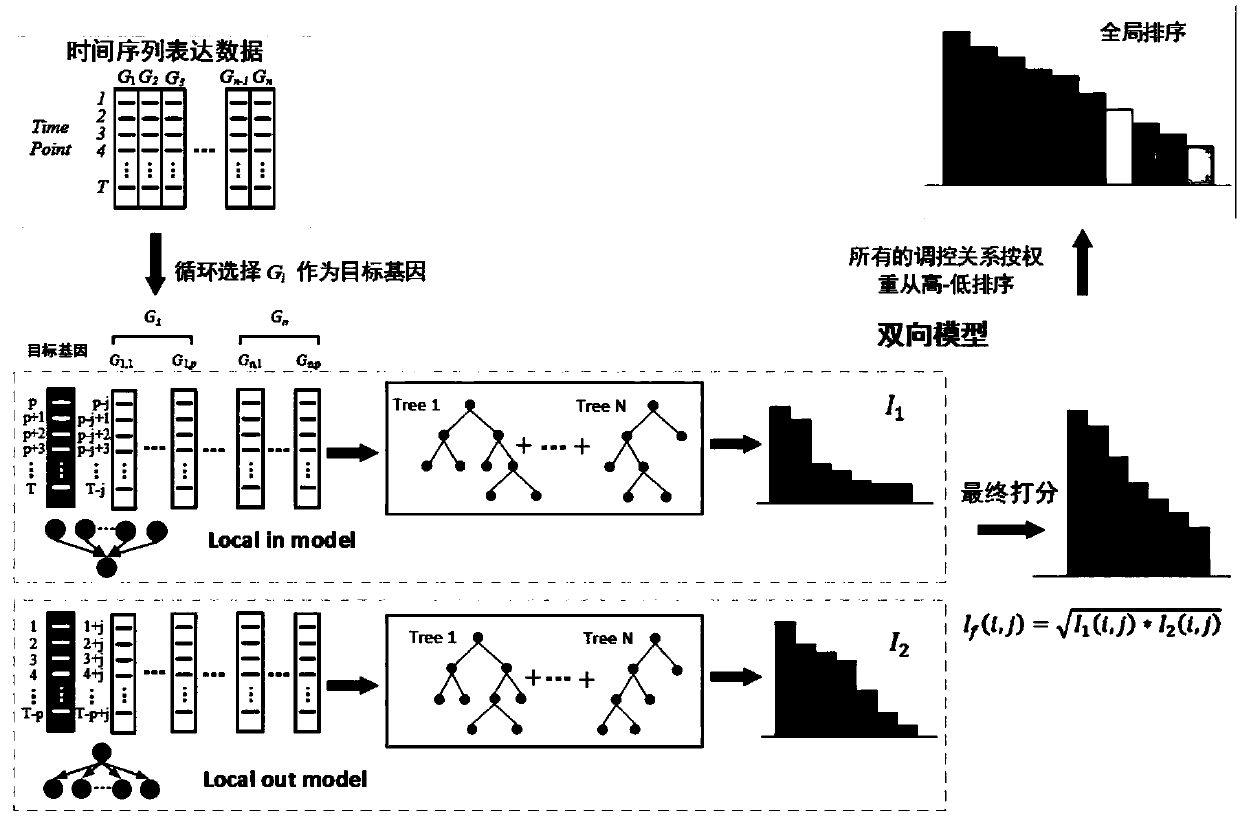
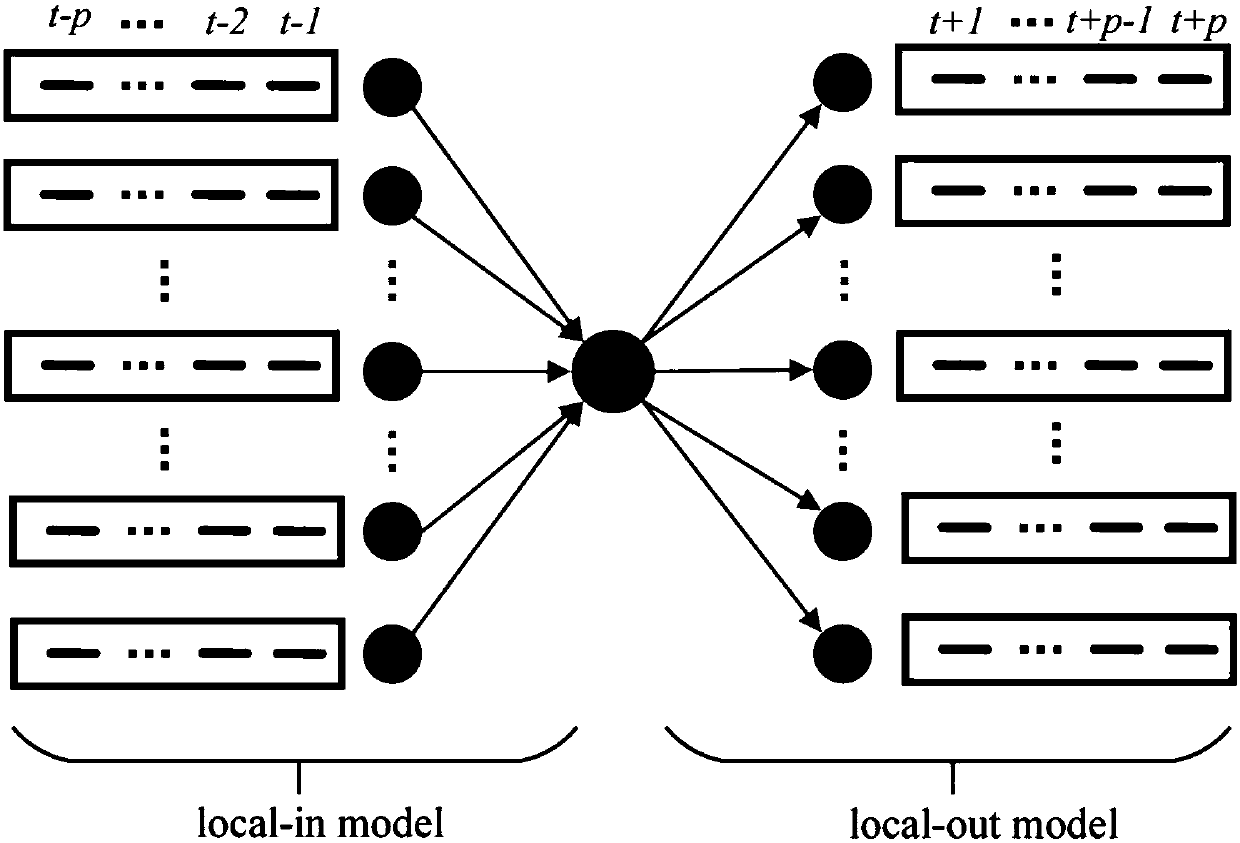
Establishment method of gene regulating network based on two-way XGBoost
The invention discloses an establishment method of a gene regulating network based on two-way XGBoost. Aiming at the characteristics of time sequence gene expression data, the relationship between anexpression value of a t moment gene and the positions before and after other gene t moment points is analyzed, and local-in and local-out two-way models are established; in a single model, establishment of the gene regulating network is divided into multiple regression equations, the condition of high-order time lag is fully considered, weights of characteristics in a single regression equation are evaluated through the XGBoost, and finally a rank of regulating relationships is acquired; normalization residuals are adopted for carrying out secondary weighting on the characteristics in each independent regression equation, and the weights of the two-way models for the same pair of regulating relationships are combined to form a value to serve as the final score of each pair of gene regulating relationships. The method is suitable for establishing different scales of gene regulating networks under the time sequence gene expression data, and the accuracy of network deduction can be effectively improved.
Owner:CENT SOUTH UNIV
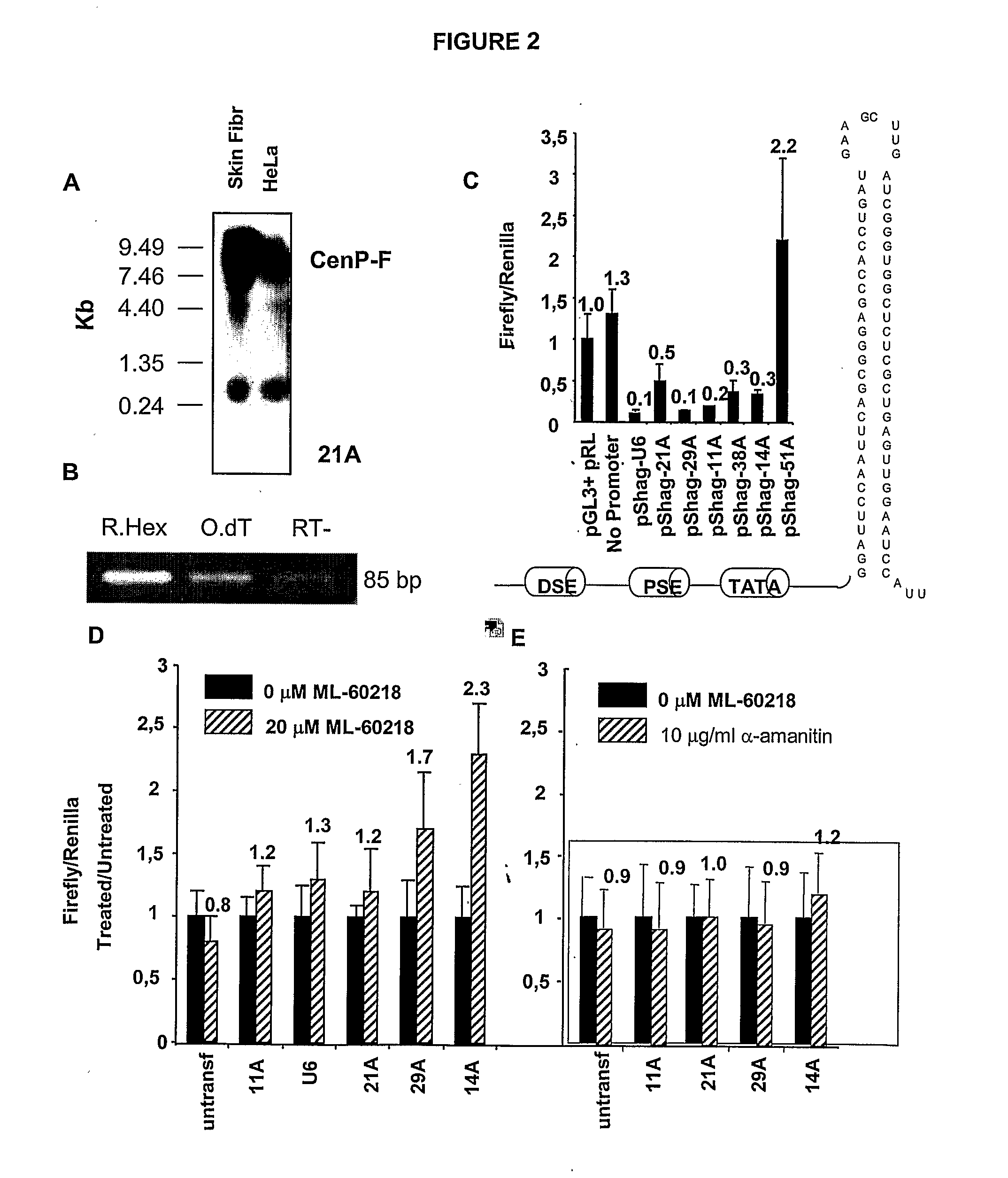
snRNA gene-like transcriptional units and uses thereof
By a computer search for upstream promoter elements (DSE, PSE) typical of small nuclear RNA (snRNA) genes, we have identified a number of previously unrecognized, putative transcription units whose predicted products are novel noncoding RNAs with homology to protein-coding genes. By elucidating the function of one of them, we provide evidence for the existence of a sense / antisense-based gene regulation network where part of the Pol III transcriptome could control its Pol II counterpart.
Owner:UNIV DEGLI STUDI DI GENOVA
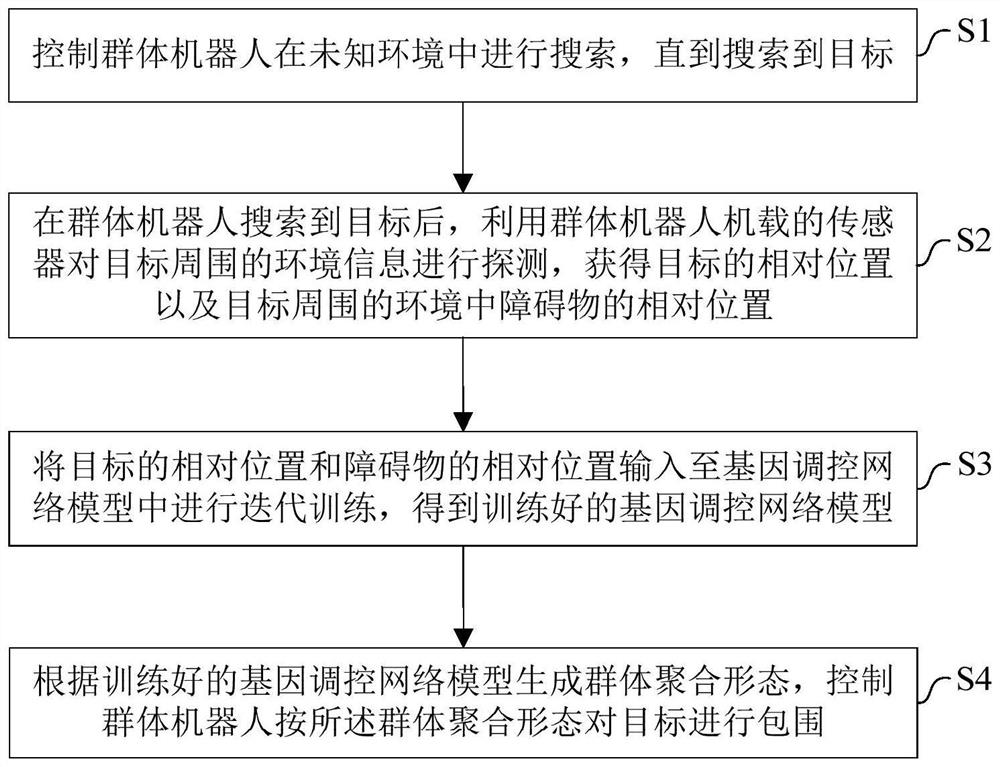
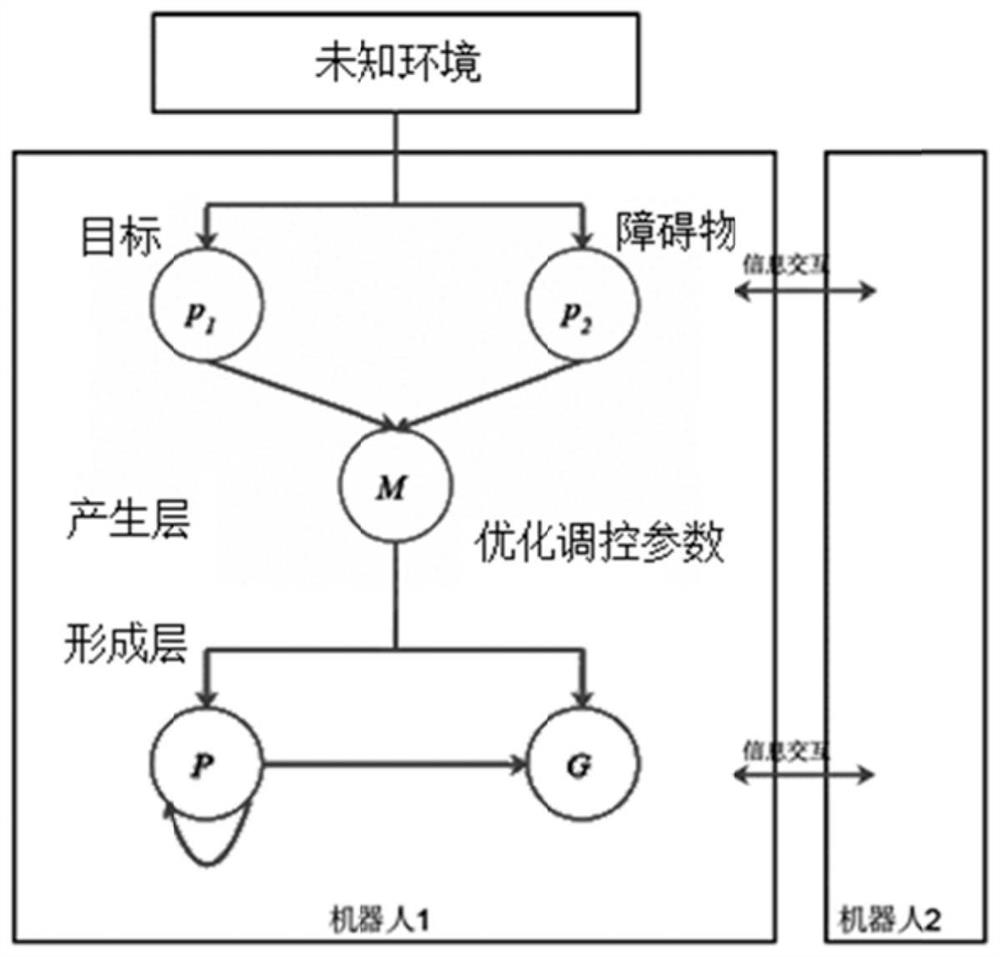
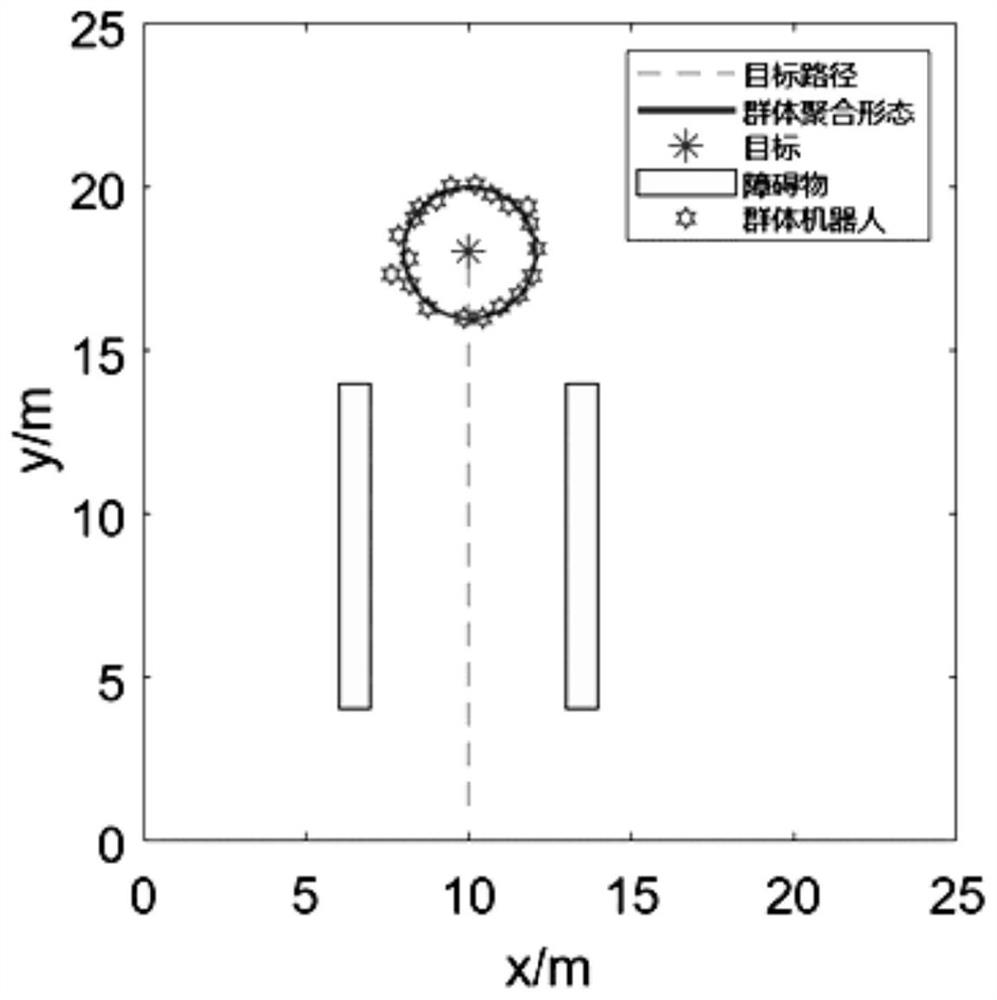
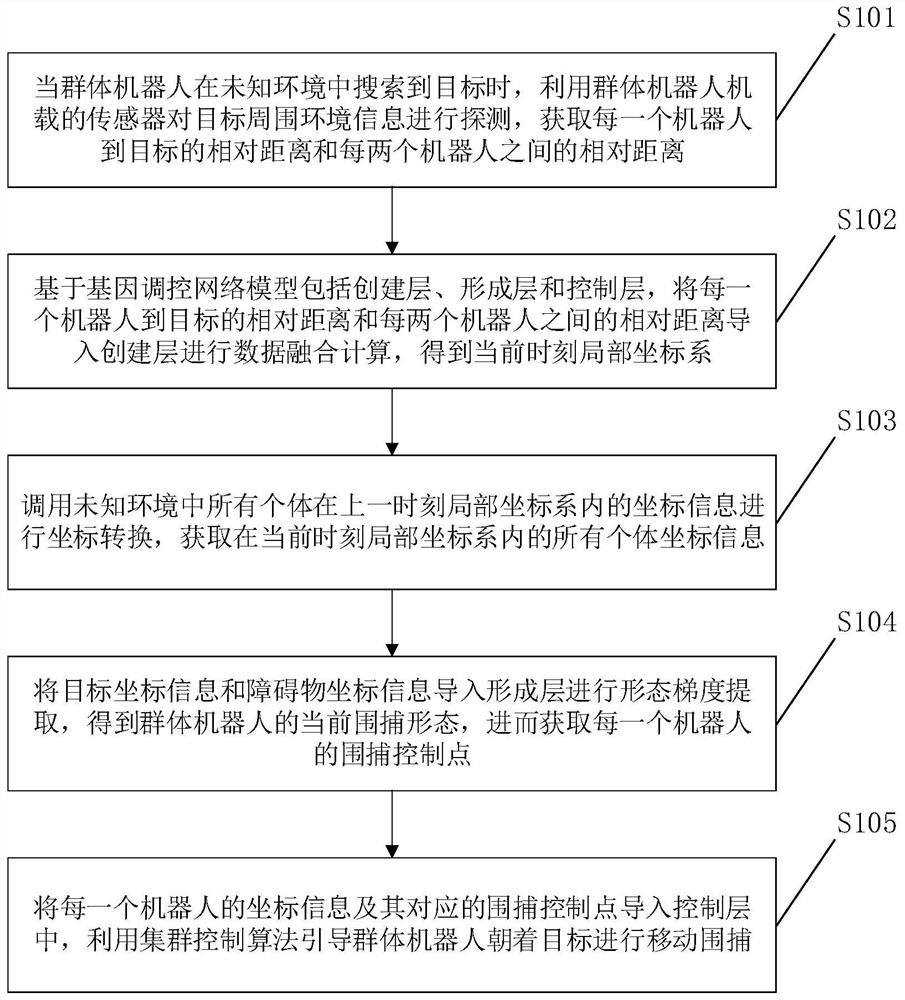
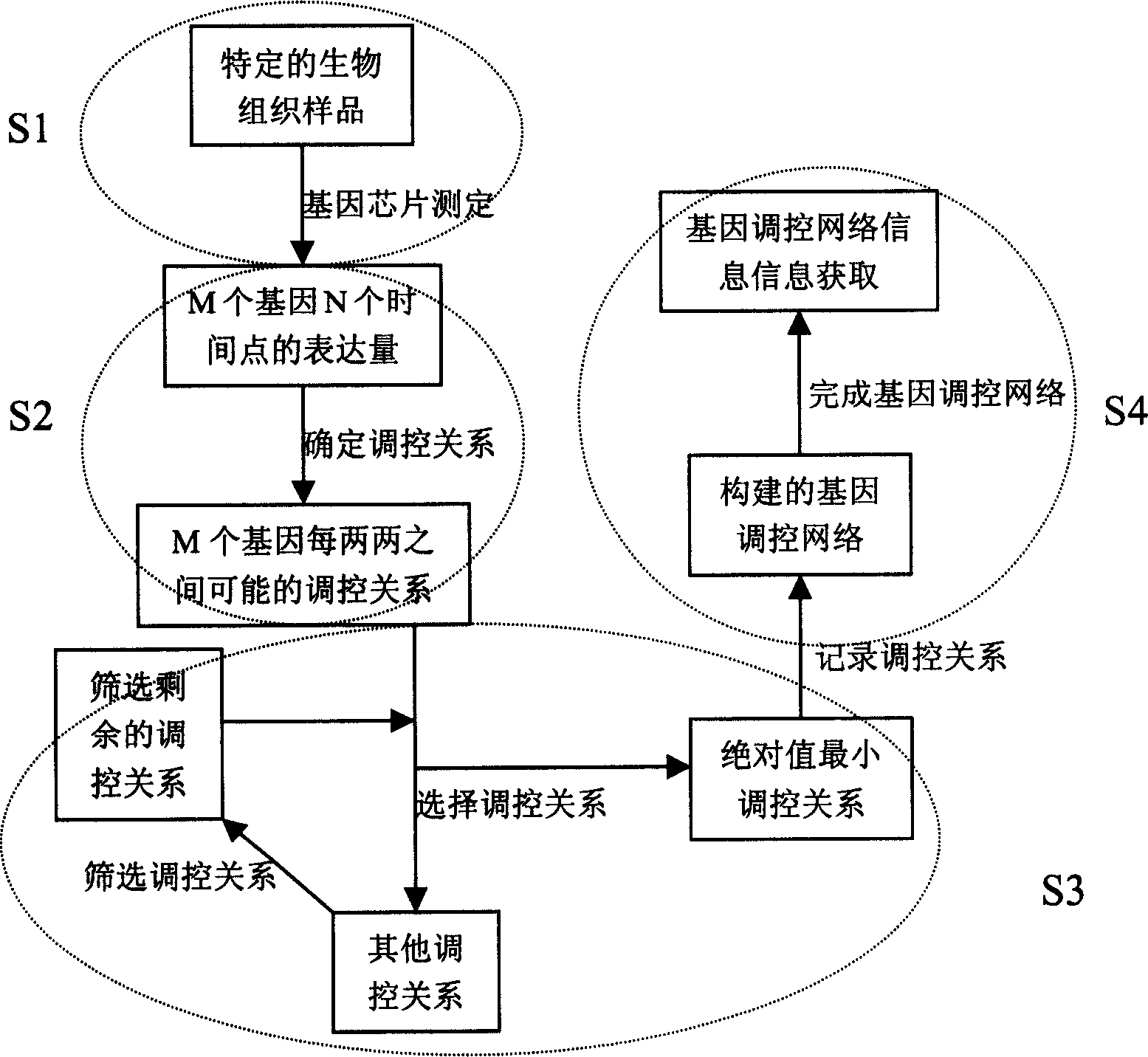
Swarm robot dynamic encirclement control method and system based on gene regulation and control network
ActiveCN112462779AAdjust Aggregate Morphology BehaviorTotal factory controlPosition/course control in two dimensionsRobot dynamicControl theory
The invention relates to the technical field of swarm intelligence, in particular to a swarm robot dynamic encirclement control method and system based on a gene regulation and control network, and the method comprises the steps: controlling swarm robots to search in an unknown environment until a target is searched; after the swarm robot searches the target, detecting the environment informationaround the target by using a sensor carried by the swarm robot to obtain the relative position of the target and the relative position of an obstacle in the environment around the target; then, inputting the relative position of the target and the relative position of the obstacle into the gene regulation and control network model for iterative training to obtain a trained gene regulation and control network model; finally, generating a swarm aggregation form according to the trained gene regulation and control network model, controlling the swarm robot to surround the target according to theswarm aggregation form. The method can efficiently complete the encirclement task in real time in a complex, dynamic and unknown environment.
Owner:SHANTOU UNIV
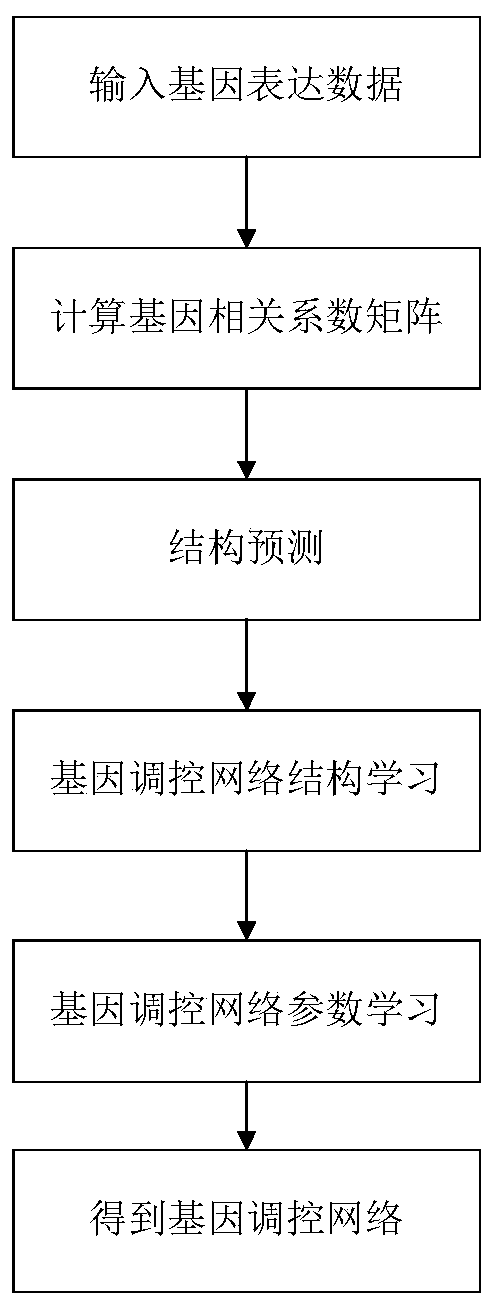
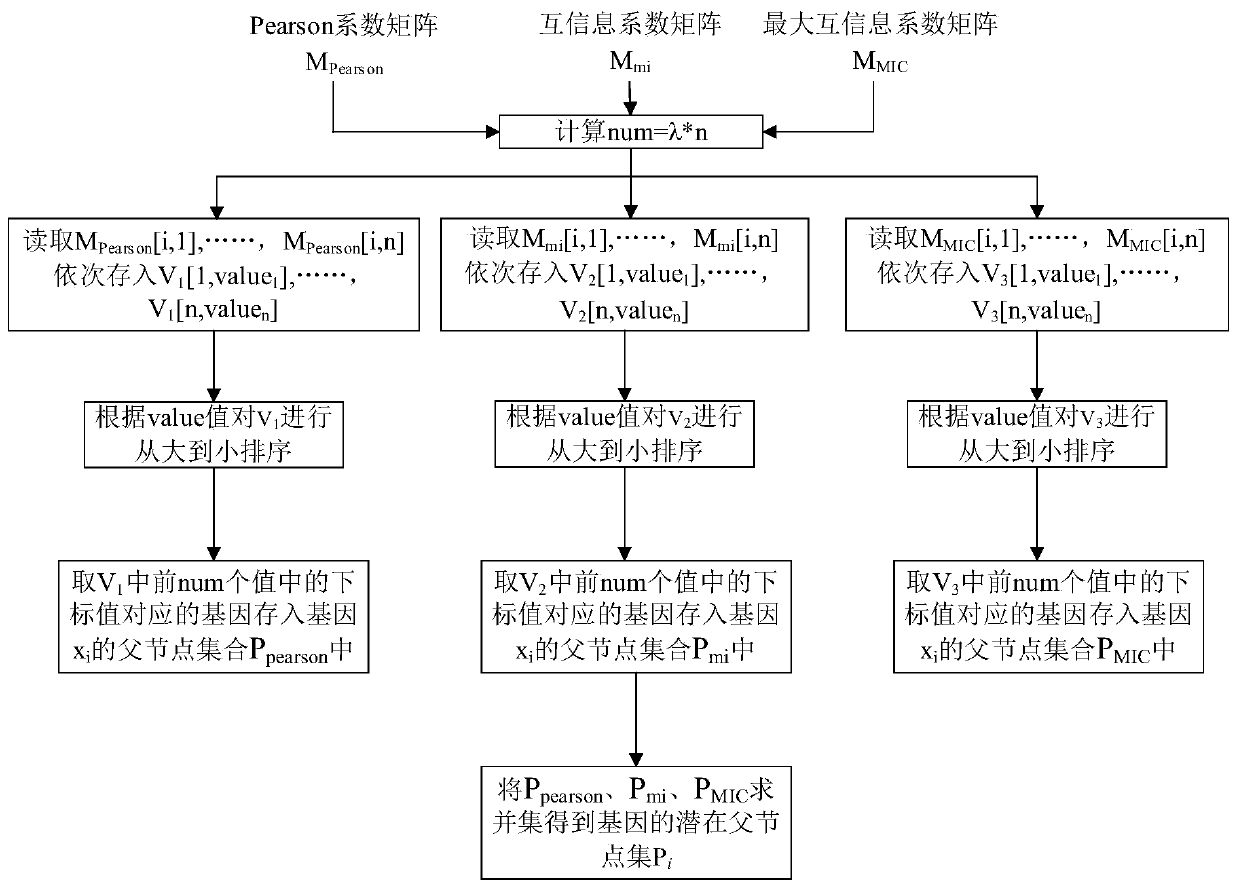
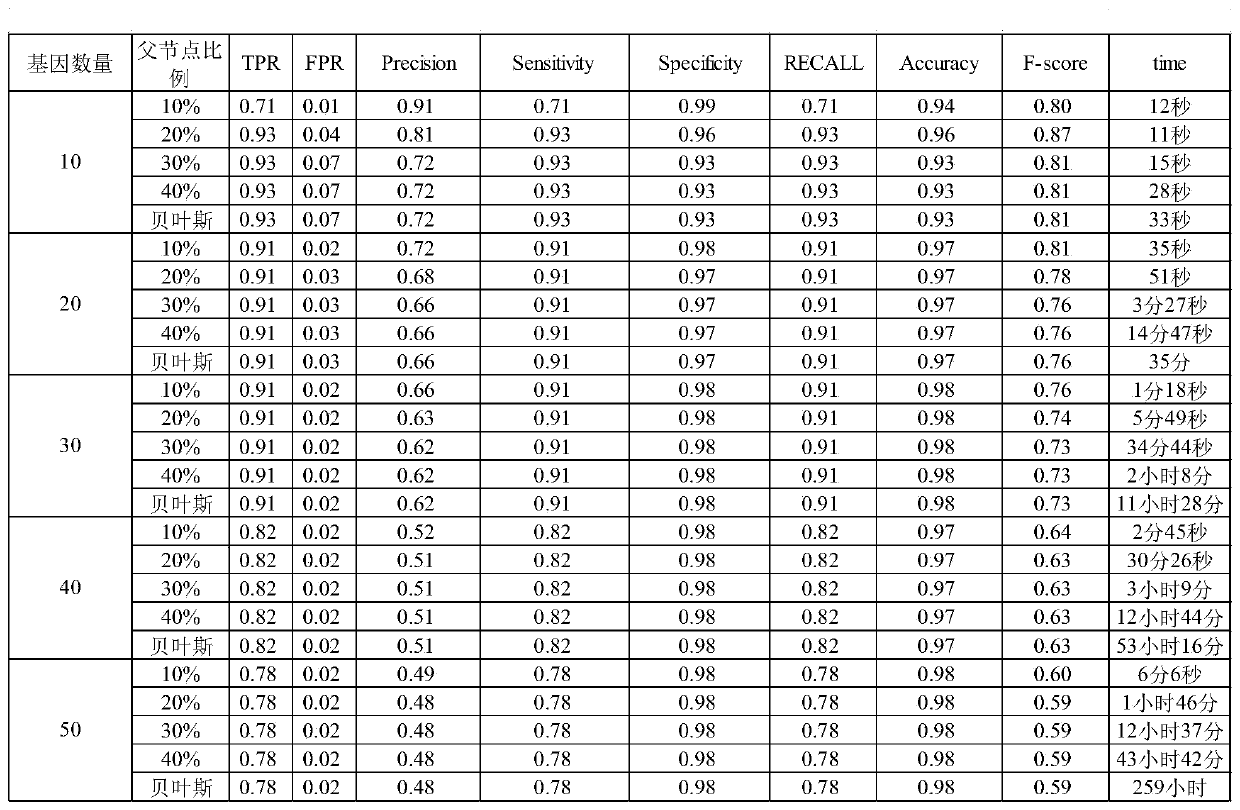
Method for constructing gene regulatory network based on structural prediction
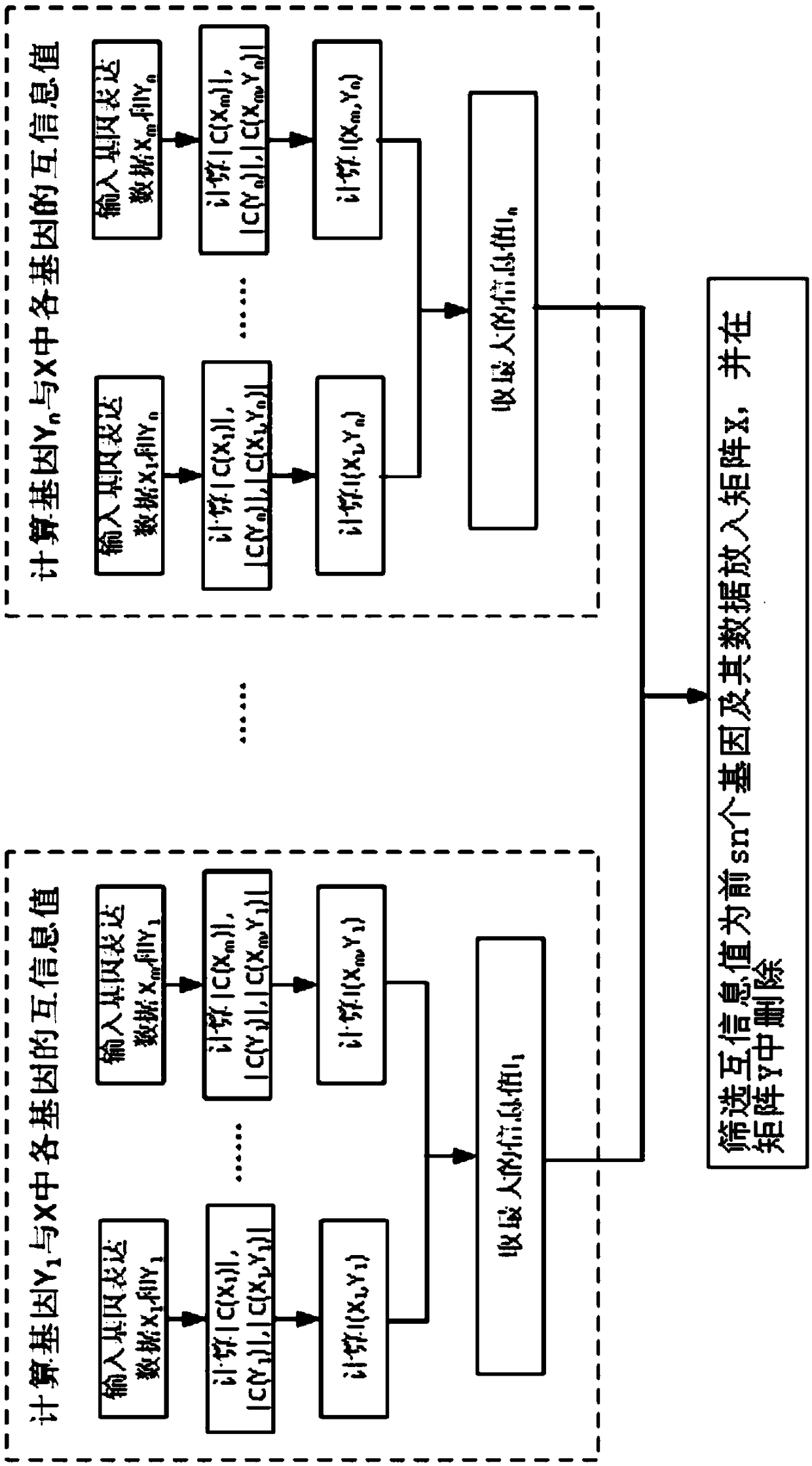
ActiveCN110675912AReduce build timeQuick buildHybridisationSystems biologyStructure learningData mining
The invention provides a method for constructing a gene regulatory network based on structural prediction. The method includes the following steps: first calculating a coefficient matrix, determiningthe correlation between genes, by calculating the Peason coefficient, the mutual information, and the maximum mutual information between the genes, as a basis for screening potential parent node sets;then, performing structural prediction, using an obtained coefficient matrix between genes as the basis for determining the potential parent node sets of the genes, and selecting a potential parent node set for each gene; finally, performing the structural learning and parameter learning of the gene regulatory network. The method predicts the potential parent node sets of the genes by a method based on of the combination of the Person coefficient, the mutual information, and the maximum mutual information, narrows the search range of structure learning, shortens the construction time of the gene regulatory network to a certain extent, improves computing performance, can build a large-scale gene regulatory network quickly and accurately, and further understands the gene regulatory mechanism of organisms.
Owner:NORTHEASTERN UNIV
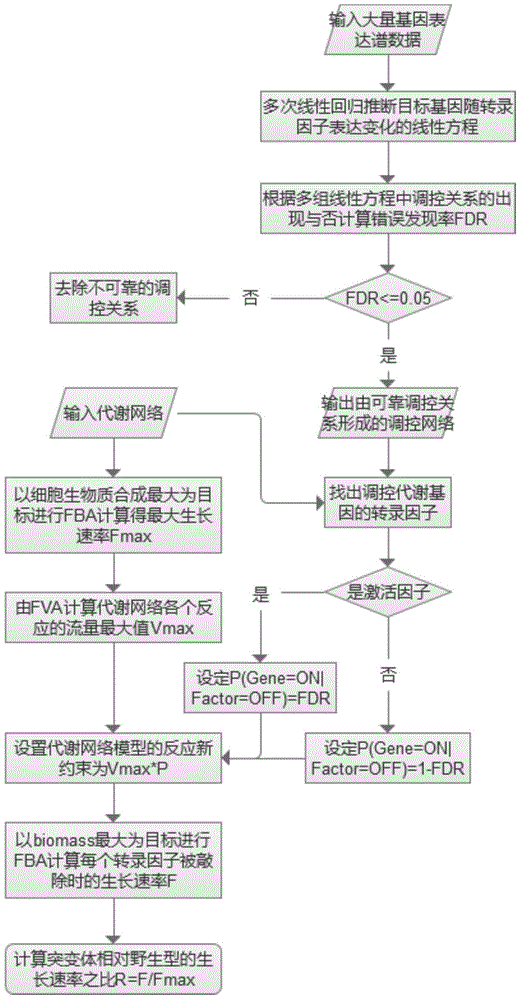
Microbial growth phenotype predication method based on control-metabolic network integration model
InactiveCN105184049AEasy to predictHigh precisionSpecial data processing applicationsMicroorganismMetabolic network
The present invention discloses a microbial growth phenotype predication method based on a control-metabolic network integration model. The method comprises: firstly, constructing a gene control network, running multiple linear regression based on a large number of gene expression profile data so as to deduce a linear equation of expression variation of each target gene with a transcription factor, and calculating a fault discovery rate (FDR); and then taking a control relation (the FDR is less than or equal to 0.05) as a global control network, finding out transcription factors of control metabolic genes in the control network, calculating a reaction flow value F corresponding to a growth rate during a knockout period of the transcription factors according to the types of the transcription factors, performing same flow equilibrium analysis by an original metabolic network so as to obtain a maximal cell growth rate Fmax, and predicating growth phenotype changes of microorganisms during the knockout period of the transcription factors by calculating F / Fmax. The method provided by the present invention enables analysis precision to be improved, so that a growth phenotype of the microorganisms can be better predicted.
Owner:SHANGHAI JIAO TONG UNIV
Group robot polymerization control method based on three-layer gene regulation and control network
ActiveCN114397887AImplement self-updatingEnsure mobile stabilityTotal factory controlPosition/course control in two dimensionsThree-dimensional spaceControl theory
The invention discloses a swarm robot aggregation control method based on a three-layer gene regulatory network, which comprises a creation layer, a formation layer and a control layer based on a gene regulatory network model, importing the relative distance from each robot to the target and the relative distance between every two robots, which are acquired by a swarm robot airborne sensor, into a creation layer to obtain a local coordinate system at the current moment; calling the coordinate information of all individuals in the local coordinate system at the previous moment to perform coordinate conversion, and acquiring target coordinate information, obstacle coordinate information and coordinate information of each robot in the local coordinate system at the current moment; the target coordinate information and the obstacle coordinate information are imported into a formation layer, a surrounding form is obtained, and a surrounding control point of each robot is obtained; and the coordinate information and the surrounding control point of each robot are imported into a control layer, and the group robots are guided to move towards the target for surrounding. According to the invention, the self-adaptive hunting of the target in the three-dimensional space with the failure of the global positioning system is realized.
Owner:SHANTOU UNIV
Process of constructing gene regulation network by continuous gene expression spectrum
InactiveCN1560271AAvoid the limitations of extracting gene regulatory networksMicrobiological testing/measurementPhase differenceChips gene expression
The invention relates to a biochip gene expression spectrum system, especially a method of constructing gene regulation and control network by continuous gene expression spectrum. It considers the size, direction and time phase difference of gene regulation and control. By large-scale gene chip continuous expression spectrum data, it considers the change condition of expression quantity with the time, calculates the distance of regulation and control relationship between genes, makes clustering analysis on the distance to construct a large-scale gene regulation and control network. The steps: a), obtaining several continuous expression spectrum gene chip data at time points of several genes; b), determining the distance of regulation and control relation between all the genes; c) selecting specific distances to construct gene regulation and control network.
Owner:INST OF COMPUTING TECH CHINESE ACAD OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com