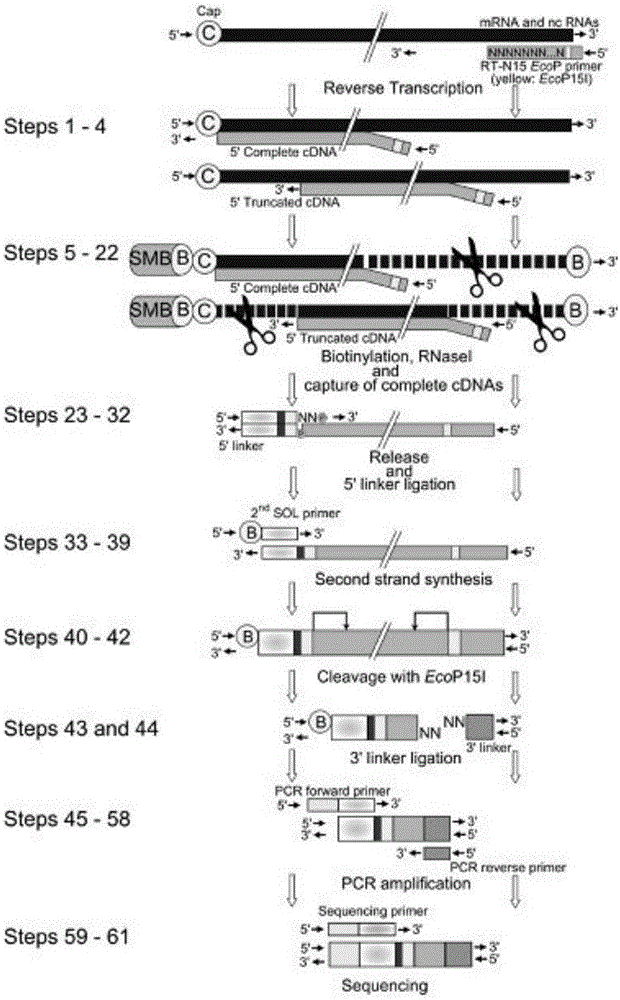
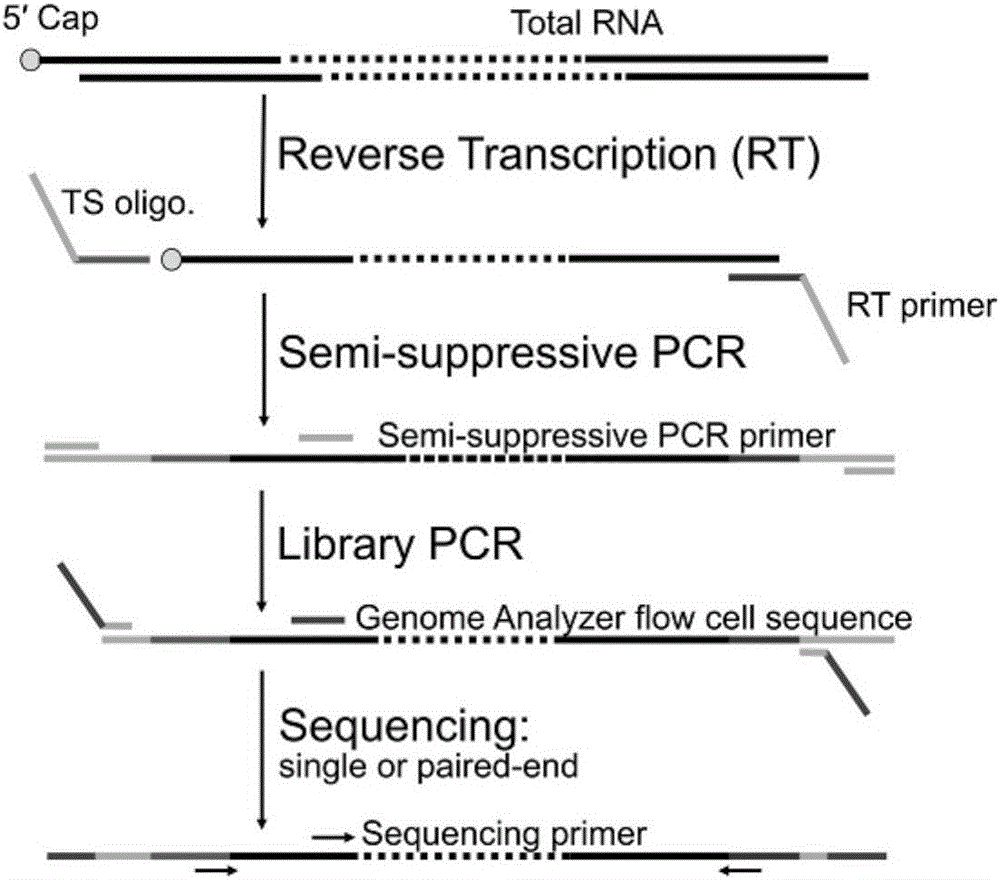
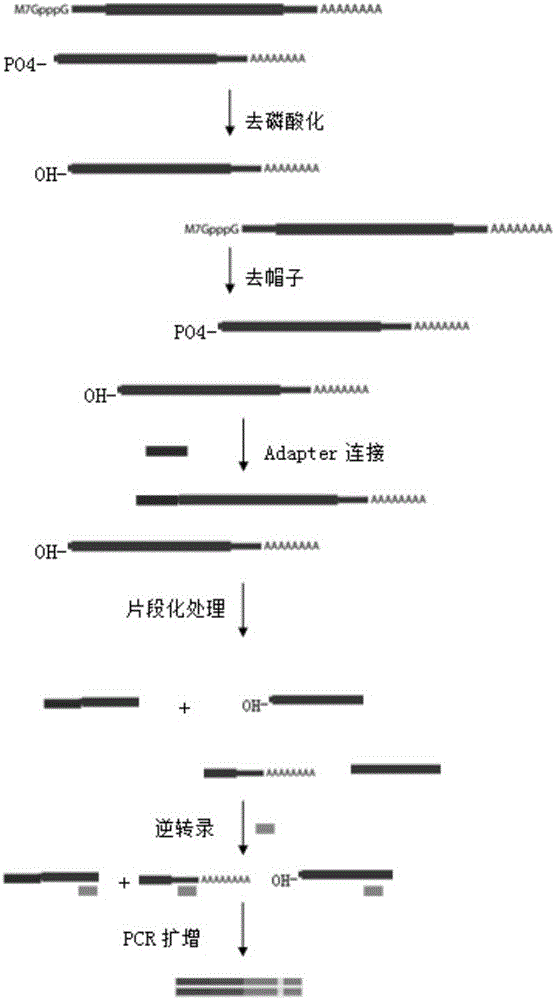
Method for constructing mRNA 5'-termnal information library
A construction method and technology for sequencing libraries, which are applied in the fields of chemical library, DNA preparation, recombinant DNA technology, etc., can solve the problems of many technical steps, no specificity, and many amplification cycles. , the effect of simplifying the construction method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] [Example 1] Capturing samples
[0040] Take 20 μg of RNA and add DNaseI enzyme (promega) for digestion to digest the DNA in the RNA sample to prevent genome contamination. Purified RNA, use 60μl Oligo(dT)25 calls up mRNA with a polyA tail. The step of purifying mRNA by magnetic beads can follow the standard procedure provided by Ambion. The sample after two rounds of elution was redissolved in 20 μl of 10 mM Tris-HCl.
[0041] Here are the detailed steps:
[0042] 1.1. Prepare the water bath and thermomixer.
[0043] 1.2. After heating the RNA sample in a water bath at 65°C for 5 minutes (to destroy the secondary structure), put it on ice quickly.
[0044] 1.3. Take 60μl Oligo(dT)25 into a new RNase-free EP tube.
[0045] 1.4. Add 100 μl BeadBindingBuffer and gently vortex to mix and stir for 1 minute, flash shake, put it on the magnetic stand, discard the supernatant, wash the beads again with BeadBindingBuffer, and remove the supernatant.
[0046] 1.5. Take 5...
Embodiment 2
[0055] [Example 2] The 5' end cap structure is connected to the 5' connector
[0056] 2.1 Dephosphorylation of RNA not protected by cap structure
[0057] First, select fastAP (NEB) enzyme, the reaction system is as follows,
[0058]
[0059] 2.2 use Oligo(dT)25 Purified Nucleic Acid
[0060] After the reaction is complete, use Oligo(dT)25 purifies nucleic acid and removes salt ions and enzymes in the reaction system. The reaction steps are as follows:
[0061] 2.2.1. Vortex the beads to mix well, take 60μl of beads and place them in a 1.5ml centrifuge tube;
[0062] 2.2.2. Take 100 μl of BindingBuffer and vortex to wash the magnetic beads, discard the supernatant for later use, repeat once, then take 50 μl of BindingBuffer to resuspend the magnetic beads;
[0063] 2.2.3. Rehydrate the sample obtained in 2.1 above to 50 μl, add it to 50 μl beads, mix by pipetting or vortexing, and incubate on a rotary mixer for 5 minutes;
[0064] 2.2.4. After incubation for 5 minut...
Embodiment 3
[0085] [Example 3] cDNA synthesis by reverse transcription
[0086] For the library construction process, refer to the RNA-seqkit operation manual of gnomegen company.
[0087] The sample was fragmented and incubated at 95°C for 5 min, and then the fragmented sample was purified with GnomeSizeSelector. Rehydrate the purified fragmented product to 40 μl, add 24 μl GnomeSizeSelector and pipette 5 times to mix, then add 16 μl 100% Ethanol, pipette 5 times to mix. Stand at room temperature for 5 minutes. Transfer the sample to a magnetic stand and let it stand for 3 minutes. Discard the supernatant. Put the sample on the magnetic stand, add 100 μl of 70% Ethanol and pipette for 3 times to wash. Absorb the 70% Ethanol cleanly, remove the sample from the magnetic stand, and let it dry for 5 minutes. Take 10 μl of DEPC water for each sample to redissolve, and pipette 5 times to resuspend the magnetic beads. Rotate the sample at a slow speed for 2 minutes, then place it on the m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com